QA: Difference between revisions
imported>Bobd |
|||
| (10 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
=Finding the QA report= | =Finding the QA report= | ||
[[File:NIMS_QA.png|thumb|QA reports appear in the datasets column of NIMS.]] | [[File:NIMS_QA.png|thumb|QA reports appear in the datasets column of NIMS.]] | ||
The QA reports take anywhere from a few minutes to an hour to compute, depending on the scan resolution and duration. To view the report, double-click the QA dataset and a pop-over will open showing the motion and spike plots, as well as some global QA values. | The QA reports take anywhere from a few minutes to an hour to compute, depending on the scan resolution and duration. To view the report, double-click the QA dataset and a pop-over will open showing the motion and spike plots, as well as some global QA values. | ||
=What's in the QA report= | =What's in the QA report= | ||
The QA code is still under active development, so we will likely be adding more metrics to the report in the near future. Right now, the report will give you | The QA code is still under active development, so we will likely be adding more metrics to the report in the near future. Right now, the report will give you the QA version number (the version of the QA script that was used to generate the report), as well as the following information: | ||
==Temporal SNR== | |||
This is the median tSNR of all brain voxels (as defined by the [http://nipy.org/dipy/examples_built/brain_extraction_dwi.html median_otsu algorithm]). The tSNR is computed after the data have been motion-corrected (details below) and the slow-drift has been removed using a 3rd-order polynomial. Note that tSNR is very sensitive to voxel size, with bigger voxels generally producing higher tSNR. The acceleration factor will also affect tSNR (both inplane acceleration and slice multiplex acceleration), with more acceleration leading to lower tSNR. It is also somewhat sensitive to the TR (with longer TRs producing slightly higher tSNR). So it's a useful metric for comparison across scans with the same voxel size, TR and acceleration, but not useful for comparing across scans with different parameters. | |||
==Spike count== | |||
The number of "spikes" detected. The spikes are detected by a simple threshold of the time series z-score plot (details below). The threshold is currently set to 6. This is somewhat arbitrary (and thus why we show the full plots), but we've found that spikes of this magnitude are generally indicative of a problem, such as excessive subject motion or scan hardware issues. | |||
==Subject Motion== | |||
Subject motion is estimated using [http://nipy.org/nipy/stable/api/generated/nipy.algorithms.registration.groupwise_registration.html FmriRealign4d] (without slice-time correction). A plot of the mean displacement, both absolute and relative, is presented. Absolute displacement is the mean displacement relative to the middle frame. Relative displacement is the mean displacement relative to the previous frame. (Mean displacement is computed using [http://www.fmrib.ox.ac.uk/analysis/techrep/tr99mj1/tr99mj1/index.html Mark Jenkinson's algorithm].) | |||
==Timeseries z-score== | |||
A plot of the mean signal (in z-score units) from each slice of the brain. This last plot is useful for detecting spikes in your data, and for determining if the spikes are caused by your subject (e.g., motion) or by a possible problem with the scanner (e.g., white-pixel noise). When a subject moves, even a little, you will often see spikes that span several or all slices. But a white-pixel noise problem typically only affects one slice at a time. Note that the first few time points are ignored for the spike plot. | |||
For this plot (as well as the motion plot) you can get the exact value of any datapoint by hovering your mouse over one of the curves. Also note that the frame numbers start at zero rather than one. Some examples of QA reports are shown below. | |||
=Artifacts that you may find= | =Artifacts that you may find= | ||
| Line 15: | Line 29: | ||
==White-pixel noise== | ==White-pixel noise== | ||
Spike noise is a common and insidious problem with MR, often caused by a loose screw on the scanner or some small stay piece of metal in the scan room that accumulates energy and then discharges randomly, creating broad-band RF noise at some point during the signal read-out. When this happens, one spot in k-space will have an abnormally high intensity and show up as a "white pixel". In the image domain, it will often manifest as an abrupt signal | Spike noise is a common and insidious problem with MR, often caused by a loose screw on the scanner or some small stay piece of metal in the scan room that accumulates energy and then discharges randomly, creating broad-band RF noise at some point during the signal read-out. When this happens, one spot in k-space will have an abnormally high intensity and show up as a "white pixel". In the image domain, it will often manifest as an abrupt signal drop in one slice at one time-point (a 'spike' in the time series). The problem is particularly acute for EPI scans because of all the gradient blipping during the read-out. | ||
If you see a lot of spike-noise in your data (either motion-induced or from a white-pixel noise problem), there are various tools available to specifically clean up spike-noise artifacts (like AFNI's 3dDespike). FSL's Melodic can also be used to remove artifacts in general (see [http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/MELODIC#fsl_regfilt_command-line_program fsl_regfit]). You can also try adding the spikes to your GLM as nuisance regressors. If you see a couple of spikes here and there, you might be able to safely ignore them, as they will not have a big effect on most GLM-type analyses. But even one or two spikes can affect certain kinds of correlation analyses, so for that you will have to be more careful. | If you see a lot of spike-noise in your data (either motion-induced or from a white-pixel noise problem), there are various tools available to specifically clean up spike-noise artifacts (like AFNI's 3dDespike). FSL's Melodic can also be used to remove artifacts in general (see [http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/MELODIC#fsl_regfilt_command-line_program fsl_regfit]). You can also try adding the spikes to your GLM as nuisance regressors. If you see a couple of spikes here and there, you might be able to safely ignore them, as they will not have a big effect on most GLM-type analyses. But even one or two spikes can affect certain kinds of correlation analyses, so for that you will have to be more careful. | ||
| Line 21: | Line 35: | ||
=Examples of QA reports= | =Examples of QA reports= | ||
A good subject and well-behaved scanner: | A good subject and well-behaved scanner: | ||
[[Image: qa_good. | [[Image: qa_good.png| 650px]] | ||
A bad subject and well-behaved scanner: | A bad subject and well-behaved scanner: | ||
[[Image: qa_motion. | [[Image: qa_motion.png| 650px]] | ||
A good subject and spikey scanner: | A good subject and spikey scanner: | ||
[[Image: qa_spikes. | [[Image: qa_spikes.png| 650px]] | ||
=Technical Details= | =Technical Details= | ||
The QA report generation code is part of the NIMS codebase and is [https://github.com/cni/nims/blob/master/nimsproc/qa_report.py available on Github]. | The QA report generation code is part of the NIMS codebase and is [https://github.com/cni/nims/blob/master/nimsproc/qa_report.py available on Github]. | ||
Latest revision as of 17:38, 23 August 2023
Quality Assurance (QA) reports are generated for all functional MR data acquired at the CNI. These reports should appear in the "datasets" column in the NIMS browser page within an hour after the scan finishes.
Finding the QA report
The QA reports take anywhere from a few minutes to an hour to compute, depending on the scan resolution and duration. To view the report, double-click the QA dataset and a pop-over will open showing the motion and spike plots, as well as some global QA values.
What's in the QA report
The QA code is still under active development, so we will likely be adding more metrics to the report in the near future. Right now, the report will give you the QA version number (the version of the QA script that was used to generate the report), as well as the following information:
Temporal SNR
This is the median tSNR of all brain voxels (as defined by the median_otsu algorithm). The tSNR is computed after the data have been motion-corrected (details below) and the slow-drift has been removed using a 3rd-order polynomial. Note that tSNR is very sensitive to voxel size, with bigger voxels generally producing higher tSNR. The acceleration factor will also affect tSNR (both inplane acceleration and slice multiplex acceleration), with more acceleration leading to lower tSNR. It is also somewhat sensitive to the TR (with longer TRs producing slightly higher tSNR). So it's a useful metric for comparison across scans with the same voxel size, TR and acceleration, but not useful for comparing across scans with different parameters.
Spike count
The number of "spikes" detected. The spikes are detected by a simple threshold of the time series z-score plot (details below). The threshold is currently set to 6. This is somewhat arbitrary (and thus why we show the full plots), but we've found that spikes of this magnitude are generally indicative of a problem, such as excessive subject motion or scan hardware issues.
Subject Motion
Subject motion is estimated using FmriRealign4d (without slice-time correction). A plot of the mean displacement, both absolute and relative, is presented. Absolute displacement is the mean displacement relative to the middle frame. Relative displacement is the mean displacement relative to the previous frame. (Mean displacement is computed using Mark Jenkinson's algorithm.)
Timeseries z-score
A plot of the mean signal (in z-score units) from each slice of the brain. This last plot is useful for detecting spikes in your data, and for determining if the spikes are caused by your subject (e.g., motion) or by a possible problem with the scanner (e.g., white-pixel noise). When a subject moves, even a little, you will often see spikes that span several or all slices. But a white-pixel noise problem typically only affects one slice at a time. Note that the first few time points are ignored for the spike plot.
For this plot (as well as the motion plot) you can get the exact value of any datapoint by hovering your mouse over one of the curves. Also note that the frame numbers start at zero rather than one. Some examples of QA reports are shown below.
Artifacts that you may find
Subject Motion
This is by far the dominant cause of spike-like artifacts in most datasets. Even a small relative head displacement can lead to a signal drop and/or increase. Motion usually affects many slices.
White-pixel noise
Spike noise is a common and insidious problem with MR, often caused by a loose screw on the scanner or some small stay piece of metal in the scan room that accumulates energy and then discharges randomly, creating broad-band RF noise at some point during the signal read-out. When this happens, one spot in k-space will have an abnormally high intensity and show up as a "white pixel". In the image domain, it will often manifest as an abrupt signal drop in one slice at one time-point (a 'spike' in the time series). The problem is particularly acute for EPI scans because of all the gradient blipping during the read-out.
If you see a lot of spike-noise in your data (either motion-induced or from a white-pixel noise problem), there are various tools available to specifically clean up spike-noise artifacts (like AFNI's 3dDespike). FSL's Melodic can also be used to remove artifacts in general (see fsl_regfit). You can also try adding the spikes to your GLM as nuisance regressors. If you see a couple of spikes here and there, you might be able to safely ignore them, as they will not have a big effect on most GLM-type analyses. But even one or two spikes can affect certain kinds of correlation analyses, so for that you will have to be more careful.
Examples of QA reports
A good subject and well-behaved scanner: File:Qa good.png
A bad subject and well-behaved scanner:
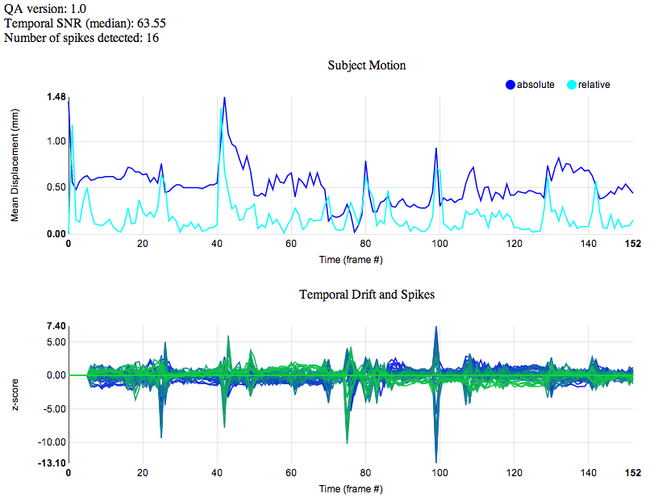
A good subject and spikey scanner:

Technical Details
The QA report generation code is part of the NIMS codebase and is available on Github.