Flywheel: Difference between revisions
imported>Wandell |
|||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
The CNI uses [//flywheel.io Flywheel] for data and computational management. Data from the scanner are automatically uploaded to the Flywheel database. The Flywheel system is on the Google Cloud Platform. Users download | The CNI uses [//flywheel.io Flywheel] for data and computational management. Data from the scanner are automatically uploaded to the Flywheel database. The Flywheel system is on the Google Cloud Platform. Users download data either using a web-browser or another of the [//docs.flywheel.io/hc/en-us/articles/360008410994-Getting-Started-Downloading-Data methods provided by Flywheel] (e.g., command-line-interface or software-development-kit). | ||
At the CNI, as data come into the Flywheel they are automatically processed according to rules determined by the CNI staff. For example, upon ingest PHI data are removed, DICOM and raw PFile data are converted to NIfTI format, and metadata about the scan parameters are read and inserted into the database. Some labs may configure initial processing of the data for individual projects differently. For help with customization groups can contact Flywheel directly or ask Michael Perry (see below). | |||
This page describes Flywheel features that are specific to the CNI. You can learn more about Flywheel using the links below: | |||
* A 5 minute video overview of Flywheel is available [//youtu.be/JUoSejTxXUw on YouTube] | |||
* Manual pages and basic material about the system are maintained by Flywheel [//docs.flywheel.io/hc/en-us at this site]. | |||
* Links to videos and webinars are on the [//docs.flywheel.io/hc/en-us Flywheel Docs site] | |||
'''Log in to CNI's Flywheel site here: [//cni.flywheel.io cni.flywheel.io]''' | |||
= Authentication = | = Authentication = | ||
Authentication to Flywheel at the CNI requires a valid (full-account) SUNetID. | Authentication to Flywheel at the CNI requires a valid (full-account) SUNetID. | ||
New users should ask the CNI staff to create an account. | New users should ask the CNI staff to create an account. | ||
= Data upload = | = Data upload = | ||
The CNI scanner produces DICOM, P-File, and Physio data. The sequence you are using along with your scan parameters will determine which of those data are generated. Regardless of the type, those data will make their way into Flywheel automatically via one of the Connectors, described next. | |||
== Connectors == | |||
At the CNI data are uploaded to Flywheel automatically via Connectors (aka Reapers). CNI has multiple Flywheel Connectors running, one for each of the types of data that must make their way into Flywheel. | |||
The connectors running at CNI are: | |||
* DICOM connector - communicates directly with the scanner's DICOM server to "discover" new data, package those data, and upload those data to Flywheel | |||
* P-File connector - packages and transfers raw P-File data from the console to Flywheel. | |||
* Physio connector - packages and transfers physio data from the console to Flywheel. This connector attempts to match the physio data to the correct series/acquisition within Flywheel, however if the scan is ended early, or if the prescribed duration is off from the actual duration, the physio data may not be transferred or it may end up in the wrong series. Please be sure to check your physio data and report any issues to CNI staff right away. | |||
== Getting your data to the right place == | |||
The Flywheel Connectors determine where to place data in the hierarchy using information entered into the MR console by the user and obtained from the DICOM or PFile data. | |||
We refer to the information provided by the user to help Flywheel understand where the data belong as the "Sort String". It is important to understand how to provide this information correctly so that your data find their way to your project without delay. | |||
If a mistake is made here, worry not, those data will still be uploaded to Flywheel, however they will go to a group and project which only CNI staff have access to. If you believe this has happened to you, please feel free to reach out to CNI staff and we will help get your data where it belongs. | |||
=== Flywheel Sort String (Good) === | === Flywheel Sort String (Good) === | ||
To make sure the data you collect is sent to the correct Flywheel project for your lab, you must enter a "Flywheel sort string" on the console. | To make sure the data you collect is sent to the correct Flywheel project for your lab, you must enter a "Flywheel sort string" on the console. | ||
Enter this string into the '''Patient ID''' field on the scanner console. | |||
The string format is: | |||
<subject_label>@<group>/<project> | <subject_label>@<group>/<project> | ||
| Line 30: | Line 53: | ||
=== Flywheel Sort String (Bad) === | === Flywheel Sort String (Bad) === | ||
If you do not enter the sort string correctly, the data will still be sent to Flywheel, but it will not be routed to the correct project. Instead, it will be assigned to the "unknown" group or "Unsorted" project, depending on what Flywheel can determine. | If you do not enter the sort string correctly, the data will still be sent to Flywheel, but it will not be routed to the correct project. Instead, it will be assigned to the "unknown" group or "Unsorted" project, depending on what Flywheel can determine. | ||
* If the data are sent to the unknown group, you must ask the Site Admins to retrieve your data. (Michael, Laima). | * If the data are sent to the unknown group, you must ask the Site Admins to retrieve your data. (Michael, Laima). | ||
* If the data are sent to the "Unsorted" project (case sensitive), you can find it and move it yourself. | * If the data are sent to the "Unsorted" project (case sensitive), you can find it and move it yourself. | ||
Thus, for a given piece of data coming in through a | |||
Thus, for a given piece of data coming in through a Connector: | |||
No Group, No Project --> group_id = "unknown", project = "Unsorted" | No Group, No Project --> group_id = "unknown", project = "Unsorted" | ||
| Line 41: | Line 66: | ||
=== Session Labels and Tags === | === Session Labels and Tags === | ||
In some cases, users want to | In some cases, users want to set the name of the session rather than have Flywheel use the default name (which is the exam number). | ||
You can control the session name/label by placing information in the ''AdditionalPatientHistory'' section on the console, like so: | |||
To set the '''session label'' using the following format (without the "<>"): | |||
label_<your_desired_session_label> | label_<your_desired_session_label> | ||
You may also '''tag a session''' to make it easier to find later. You can '''set a session tag''' by inserting this string in the ''AdditionalPatientHistory'' section. | |||
tag_<tag_name> | tag_<tag_name> | ||
| Line 55: | Line 85: | ||
Protected Health Information (PHI) is considered High Risk Data according to the Stanford Data Classification Guidelines. PHI is defined as any information that can be used to identify an individual and may relate to their past, present, or future health. By law, this information must be encrypted by law and must be (a) stored in encrypted form, and (b) transmitted only through secure means. | Protected Health Information (PHI) is considered High Risk Data according to the Stanford Data Classification Guidelines. PHI is defined as any information that can be used to identify an individual and may relate to their past, present, or future health. By law, this information must be encrypted by law and must be (a) stored in encrypted form, and (b) transmitted only through secure means. | ||
Anonymized research data for publication can be shared without harm. See [https://med.stanford.edu/irt/security/stanfordinfo/hipaa.html this Stanford site]. Although CNI participants are not medical patients, we treat human subject data with the same PHI status as if they were patients. Specifically, data uploaded to Flywheel strips information from these fields: | |||
* Patient ID field | |||
* Date of Birth | |||
* Medical Record Number | |||
* Subject Name (first and last) | |||
For PIs, please note we do look for PHI information and over the years we have found that users sometimes insert names, phone numbers and other information that might identify the subject. When we find such information, we delete it. The most likely places that RAs and others insert this information are these fields: | |||
* Subject code (part of Patient ID) | * Subject code (part of Patient ID) | ||
| Line 73: | Line 98: | ||
* Series description | * Series description | ||
''' | Thus, we ask all CNI users: '''Never use any PHI identifiers when entering data into the scanner!''' | ||
= Data Processing | It is permissible to include the subject's weight and age up to some precision. | ||
At present, to describe the PHI methods in your publications, you might find this summary paragraph helpful. | |||
''Data at Stanford's Center for Cognitive and Neurobiological Imaging are securely transferred from the MR scanner directly to a data management system (Flywheel.io) that is running within a Google platform space that is approved for research data. Prior to transfer the data file headers are stripped of fields that may contain subject information (patient id, DOB, MRN, name fields). These procedures meet the Stanford standard for anonymized research data for publication and can be shared without harm. See: [https://med.stanford.edu/irt/security/stanfordinfo/hipaa.html this Stanford site]. | |||
'''N.B.''' Flywheel is planning to provide a substantial enhancement of the anonymization protocol so that each lab will have the ability to control the details of its own anonymization protocol. We will send a note describing the capabilities when the feature is released - which is expected first quarter 2021. | |||
= Data Processing = | |||
Once data are uploaded to Flywheel those data are automatically processed according to a given Project's "[//docs.flywheel.io/hc/en-us/articles/360008553133-Project-Gear-Rules Gear Rules]". The Rules define which Gear will be run on a given piece of data when it lands in Flywheel. These rules do everything from metadata extraction from DICOM headers to execution of QA algorithms. | |||
== Gear Rules (Default) == | |||
At CNI we have defined a default set of rules that are applied to every new project. Each rule defines the logic that must be satisfied in order for a given gear to be run on a data file. | |||
The image below shows the default rules that every new project is provided with. | |||
[[File:GearRules.png|600px|thumb|center|Gear Rule Template]] | |||
=== A special note on DICOM to NIfTI Conversion (dcm2niix & dcm-convert) === | |||
Historically at CNI we have provided a tool that was developed in-house for DICOM to NIfTI conversion (dcm-convert). Recently users have expressed interest in having a very widely used and popular tool, dcm2niix, serve as the default tool to perform DICOM to NIfTI conversion in Flywheel. We are happy to say that Flywheel does provide a dcm2niix Gear and we have incorporated this into our default rule set. As of today the default is still dcm-convert, however for projects created recently you will see this rule available and in a disabled state. You can easily enable this rule, however it should be noted that if you do enable this rule you need to disable the existing dcm-convert rule so that you don't introduce a race condition which would result in a situation where you would not be able to predict which tool would be used to generate the NIfTI data. | |||
=== Re-running DICOM to NIfTI conversion across a project === | |||
There are special considerations to be made if you desire to re-run DICOM to NIfTI conversion across an entire project, namely you need to ignore DICOM data that was generated by any of the MUX sequences, as those data are aliased and running either of the DICOM to NIfTI tools on those data will result in aliased NIfTI data. Special care is taken within the Gear rules to avoid this situation by specifically ignoring data that were collected with a mux sequence. | |||
The best idea here is to coordinate with CNI staff (e.g., Michael Perry) prior to doing this. | |||
= Downloading Data = | = Downloading Data = | ||
There are several ways to download data from Flywheel, including via the [ | There are several ways to download data from Flywheel, including via the [//flywheelio.zendesk.com/hc/en-us/articles/360008622994-Downloading-sessions-or-acquisitions-from-the-Web-UI WEB UI], the [//flywheelio.zendesk.com/hc/en-us/sections/360001596834-Command-Line-Interface command line interface (CLI)], and the Flywheel SDK (which is available for both [//flywheel-io.github.io/core/branches/master/python/getting_started.html Python] and [//flywheel-io.github.io/core/branches/master/matlab/getting_started.html MATLAB]). The links below can help you get started with the various methods of export: | ||
=== UI Downloads === | === UI Downloads === | ||
* [//flywheelio.zendesk.com/hc/en-us/articles/360009256973-Downloading-a-file Download individual Files] | |||
* [//flywheelio.zendesk.com/hc/en-us/articles/360008622994-Downloading-sessions-or-acquisitions-from-the-Web-UI Download session or acquisition containers] | |||
* [//flywheelio.zendesk.com/hc/en-us/articles/360008629794-Downloading-an-entire-project Download a Project] | |||
=== Flywheel SDK === | === Flywheel SDK === | ||
* [//flywheel-io.gitlab.io/product/backend/sdk/branches/master/matlab/index.html MATLAB] | |||
* [//flywheel-io.gitlab.io/product/backend/sdk/branches/master/python/index.html Python] | |||
* [//gitlab.com/flywheel-io/public/flywheel-tutorials Support materials SDK examples (Jupyter notebooks)] | |||
=== Flywheel CLI === | |||
* [//docs.flywheel.io/hc/en-us/search?utf8=%E2%9C%93&query=CLI Flywheel CLI Documentation] | |||
=== CLI: Tips and Best Practices for CNI Users === | |||
== | This section focuses on a few tips for using the CLI to download your data. For a complete overview of the Flywheel CLI, including how to get started, please look through the [//docs.flywheel.io/hc/en-us/search?utf8=%E2%9C%93&query=CLI Flywheel CLI Documentation]. | ||
''Tip: Download only the files you need'' | ''Tip:'' ''' Download only the files you need''' | ||
When downloading data from Flywheel using the CLI you can greatly speed up your downloads by excluding data types which are not needed for your analysis. | When downloading data from Flywheel using the CLI you can greatly speed up your downloads by excluding data types which are not needed for your analysis. | ||
''Example: '''''Exclude pfile and DICOM data from a container download''': Most users do not need to download the raw scanner files (PFILES) or raw DICOM data. You can exclude certain data types from your downloads by using the `-e` flag with your CLI download, like so: | ''Example: '''''Exclude pfile and DICOM data from a container download''': Most users do not need to download the raw scanner files (PFILES) or raw DICOM data. You can exclude certain data types from your downloads by using the `-e` flag with your CLI download, like so: | ||
fw download "cni/testproject/subject1/session1" -e pfile -e dicom | fw download "cni/testproject/subject1/session1" -e pfile -e dicom | ||
This tells the CLI to exclude any pfile and dicom files in the container. Note that you can use consecutive -e flags to exclude multiple data types. | This tells the CLI to exclude any pfile and dicom files in the container. Note that you can use consecutive -e flags to exclude multiple data types. | ||
''Example:'' '''Download only NIfTI, BVEC, and BVAL files''':<br> | ''Example:'' '''Download only NIfTI, BVEC, and BVAL files''':<br> | ||
Most users are only interested in the data that will be input to their analysis pipelines. This is most often limited to three data types (nifti, bvec, and bval). You can use the following command with multiple include flags (`-i`) to accomplish exactly that: | Most users are only interested in the data that will be input to their analysis pipelines. This is most often limited to three data types (nifti, bvec, and bval). You can use the following command with multiple include flags (`-i`) to accomplish exactly that: | ||
fw download "cni/testproject/subject1/session1" -i nifti -i bvec -i bval | fw download "cni/testproject/subject1/session1" -i nifti -i bvec -i bval | ||
This will generate an archive (.tar) file containing the requested hierarchy with only those files you explicitly need. | This will generate an archive (.tar) file containing the requested hierarchy with only those files you explicitly need. | ||
'' | |||
''Example:'' '''Using quotes''' | |||
Often times your source-path (that is the ''group/project/subject/session'' string the describes the location of your data in Flywheel) will have one or more spaces or special characters in it. To properly address that location using the CLI it's important to use quotes around the source path, like so: | Often times your source-path (that is the ''group/project/subject/session'' string the describes the location of your data in Flywheel) will have one or more spaces or special characters in it. To properly address that location using the CLI it's important to use quotes around the source path, like so: | ||
fw download "cni/testproject/subject1/session1" | fw download "cni/testproject/subject1/session1" | ||
''Example'' ''Download a single file with the CLI'' | ''Example'' ''Download a single file with the CLI'' | ||
So you have navigated to a container and your only desire is to download a single file from that container, the best way to do that is using the CLI with the 'files' spec filter. | So you have navigated to a container and your only desire is to download a single file from that container, the best way to do that is using the CLI with the 'files' spec filter. | ||
''Example:'' '''Download a single NIfTI file from an acquisition container:''' | ''Example:'' '''Download a single NIfTI file from an acquisition container:''' | ||
fw download "test/Unsorted/s001/18591/T1w 1mm/files/18591_13_1.nii.gz" | fw download "test/Unsorted/s001/18591/T1w 1mm/files/18591_13_1.nii.gz" | ||
The important bit here is the inclusion of "files" prior to the file name. | |||
''The important bit here is the inclusion of "files" prior to the file name.'' | |||
= Using Sherlock and other Computers = | = Using Sherlock and other Computers = | ||
We suggest that you use the Flywheel Command Line Interface (CLI) to transfer data to other compute resources, like Sherlock. | |||
To download and install the CLI on Sherlock we use the `wget` tool, then `unzip` to extract the CLI resource package, and finally modify our `.bashrc` file to add the fw binary as an alias in our environment. | |||
'''Step 1:''' Get the URL for the CLI package, which can be found on the "Profile" page within the Flywheel interface. To grab the URL, find the CLI section on the Profile page, right-click the Linux CLI Download link, and choose "Copy Link Address" (or similar). Once you have the download link address move to the next step. | '''Step 1:''' Get the URL for the CLI package, which can be found on the "Profile" page within the Flywheel interface. To grab the URL, find the CLI section on the Profile page, right-click the Linux CLI Download link, and choose "Copy Link Address" (or similar). Once you have the download link address move to the next step. | ||
| Line 129: | Line 202: | ||
mkdir -p flywheel/cli | mkdir -p flywheel/cli | ||
cd flywheel/cli | cd flywheel/cli | ||
wget | wget //storage.googleapis.com/flywheel-dist/cli/<version>/fw-linux_amd64.zip | ||
| Line 144: | Line 217: | ||
'''Step 5:''' Once the above steps are complete, you should be able to log in using the CLI and use it as described in the official documentation | '''Step 5:''' Once the above steps are complete, you should be able to log in using the CLI and use it as described in the official documentation: //flywheelio.zendesk.com/hc/en-us/sections/360001596834-Command-Line-Interface. The best way to do this is navigate to your [//cni.flywheel.io/#/profile profile page] in Flywheel, make sure that you have generated an API Key, and use the login command text that is provided for you there. | ||
fw login <your API key> | fw login <your API key> | ||
Latest revision as of 17:38, 23 August 2023
The CNI uses Flywheel for data and computational management. Data from the scanner are automatically uploaded to the Flywheel database. The Flywheel system is on the Google Cloud Platform. Users download data either using a web-browser or another of the methods provided by Flywheel (e.g., command-line-interface or software-development-kit).
At the CNI, as data come into the Flywheel they are automatically processed according to rules determined by the CNI staff. For example, upon ingest PHI data are removed, DICOM and raw PFile data are converted to NIfTI format, and metadata about the scan parameters are read and inserted into the database. Some labs may configure initial processing of the data for individual projects differently. For help with customization groups can contact Flywheel directly or ask Michael Perry (see below).
This page describes Flywheel features that are specific to the CNI. You can learn more about Flywheel using the links below:
- A 5 minute video overview of Flywheel is available on YouTube
- Manual pages and basic material about the system are maintained by Flywheel at this site.
- Links to videos and webinars are on the Flywheel Docs site
Log in to CNI's Flywheel site here: cni.flywheel.io
Authentication
Authentication to Flywheel at the CNI requires a valid (full-account) SUNetID.
New users should ask the CNI staff to create an account.
Data upload
The CNI scanner produces DICOM, P-File, and Physio data. The sequence you are using along with your scan parameters will determine which of those data are generated. Regardless of the type, those data will make their way into Flywheel automatically via one of the Connectors, described next.
Connectors
At the CNI data are uploaded to Flywheel automatically via Connectors (aka Reapers). CNI has multiple Flywheel Connectors running, one for each of the types of data that must make their way into Flywheel.
The connectors running at CNI are:
- DICOM connector - communicates directly with the scanner's DICOM server to "discover" new data, package those data, and upload those data to Flywheel
- P-File connector - packages and transfers raw P-File data from the console to Flywheel.
- Physio connector - packages and transfers physio data from the console to Flywheel. This connector attempts to match the physio data to the correct series/acquisition within Flywheel, however if the scan is ended early, or if the prescribed duration is off from the actual duration, the physio data may not be transferred or it may end up in the wrong series. Please be sure to check your physio data and report any issues to CNI staff right away.
Getting your data to the right place
The Flywheel Connectors determine where to place data in the hierarchy using information entered into the MR console by the user and obtained from the DICOM or PFile data.
We refer to the information provided by the user to help Flywheel understand where the data belong as the "Sort String". It is important to understand how to provide this information correctly so that your data find their way to your project without delay.
If a mistake is made here, worry not, those data will still be uploaded to Flywheel, however they will go to a group and project which only CNI staff have access to. If you believe this has happened to you, please feel free to reach out to CNI staff and we will help get your data where it belongs.
Flywheel Sort String (Good)
To make sure the data you collect is sent to the correct Flywheel project for your lab, you must enter a "Flywheel sort string" on the console.
Enter this string into the Patient ID field on the scanner console.
The string format is:
<subject_label>@<group>/<project>
Flywheel Sort String (Bad)
If you do not enter the sort string correctly, the data will still be sent to Flywheel, but it will not be routed to the correct project. Instead, it will be assigned to the "unknown" group or "Unsorted" project, depending on what Flywheel can determine.
- If the data are sent to the unknown group, you must ask the Site Admins to retrieve your data. (Michael, Laima).
- If the data are sent to the "Unsorted" project (case sensitive), you can find it and move it yourself.
Thus, for a given piece of data coming in through a Connector:
No Group, No Project --> group_id = "unknown", project = "Unsorted" No Group, Project --> group_id = "unknown", project = <Project> Group, No Project --> group_id = <Group>, project = "Unsorted"
Session Labels and Tags
In some cases, users want to set the name of the session rather than have Flywheel use the default name (which is the exam number).
You can control the session name/label by placing information in the AdditionalPatientHistory section on the console, like so:
To set the 'session label using the following format (without the "<>"):
label_<your_desired_session_label>
You may also tag a session to make it easier to find later. You can set a session tag by inserting this string in the AdditionalPatientHistory section.
tag_<tag_name>
Protected Health Information
Protected Health Information (PHI) is considered High Risk Data according to the Stanford Data Classification Guidelines. PHI is defined as any information that can be used to identify an individual and may relate to their past, present, or future health. By law, this information must be encrypted by law and must be (a) stored in encrypted form, and (b) transmitted only through secure means.
Anonymized research data for publication can be shared without harm. See this Stanford site. Although CNI participants are not medical patients, we treat human subject data with the same PHI status as if they were patients. Specifically, data uploaded to Flywheel strips information from these fields:
- Patient ID field
- Date of Birth
- Medical Record Number
- Subject Name (first and last)
For PIs, please note we do look for PHI information and over the years we have found that users sometimes insert names, phone numbers and other information that might identify the subject. When we find such information, we delete it. The most likely places that RAs and others insert this information are these fields:
- Subject code (part of Patient ID)
- Exam description
- Series description
Thus, we ask all CNI users: Never use any PHI identifiers when entering data into the scanner!
It is permissible to include the subject's weight and age up to some precision.
At present, to describe the PHI methods in your publications, you might find this summary paragraph helpful.
Data at Stanford's Center for Cognitive and Neurobiological Imaging are securely transferred from the MR scanner directly to a data management system (Flywheel.io) that is running within a Google platform space that is approved for research data. Prior to transfer the data file headers are stripped of fields that may contain subject information (patient id, DOB, MRN, name fields). These procedures meet the Stanford standard for anonymized research data for publication and can be shared without harm. See: this Stanford site.
N.B. Flywheel is planning to provide a substantial enhancement of the anonymization protocol so that each lab will have the ability to control the details of its own anonymization protocol. We will send a note describing the capabilities when the feature is released - which is expected first quarter 2021.
Data Processing
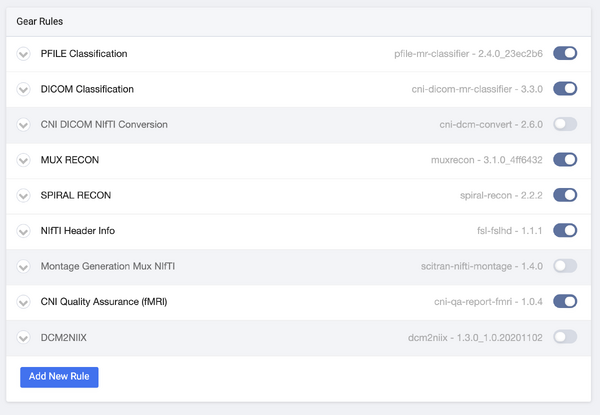
Once data are uploaded to Flywheel those data are automatically processed according to a given Project's "Gear Rules". The Rules define which Gear will be run on a given piece of data when it lands in Flywheel. These rules do everything from metadata extraction from DICOM headers to execution of QA algorithms.
Gear Rules (Default)
At CNI we have defined a default set of rules that are applied to every new project. Each rule defines the logic that must be satisfied in order for a given gear to be run on a data file.
The image below shows the default rules that every new project is provided with.
A special note on DICOM to NIfTI Conversion (dcm2niix & dcm-convert)
Historically at CNI we have provided a tool that was developed in-house for DICOM to NIfTI conversion (dcm-convert). Recently users have expressed interest in having a very widely used and popular tool, dcm2niix, serve as the default tool to perform DICOM to NIfTI conversion in Flywheel. We are happy to say that Flywheel does provide a dcm2niix Gear and we have incorporated this into our default rule set. As of today the default is still dcm-convert, however for projects created recently you will see this rule available and in a disabled state. You can easily enable this rule, however it should be noted that if you do enable this rule you need to disable the existing dcm-convert rule so that you don't introduce a race condition which would result in a situation where you would not be able to predict which tool would be used to generate the NIfTI data.
Re-running DICOM to NIfTI conversion across a project
There are special considerations to be made if you desire to re-run DICOM to NIfTI conversion across an entire project, namely you need to ignore DICOM data that was generated by any of the MUX sequences, as those data are aliased and running either of the DICOM to NIfTI tools on those data will result in aliased NIfTI data. Special care is taken within the Gear rules to avoid this situation by specifically ignoring data that were collected with a mux sequence.
The best idea here is to coordinate with CNI staff (e.g., Michael Perry) prior to doing this.
Downloading Data
There are several ways to download data from Flywheel, including via the WEB UI, the command line interface (CLI), and the Flywheel SDK (which is available for both Python and MATLAB). The links below can help you get started with the various methods of export:
UI Downloads
Flywheel SDK
Flywheel CLI
CLI: Tips and Best Practices for CNI Users
This section focuses on a few tips for using the CLI to download your data. For a complete overview of the Flywheel CLI, including how to get started, please look through the Flywheel CLI Documentation.
Tip: Download only the files you need
When downloading data from Flywheel using the CLI you can greatly speed up your downloads by excluding data types which are not needed for your analysis.
Example: Exclude pfile and DICOM data from a container download: Most users do not need to download the raw scanner files (PFILES) or raw DICOM data. You can exclude certain data types from your downloads by using the `-e` flag with your CLI download, like so:
fw download "cni/testproject/subject1/session1" -e pfile -e dicom
This tells the CLI to exclude any pfile and dicom files in the container. Note that you can use consecutive -e flags to exclude multiple data types.
Example: Download only NIfTI, BVEC, and BVAL files:
Most users are only interested in the data that will be input to their analysis pipelines. This is most often limited to three data types (nifti, bvec, and bval). You can use the following command with multiple include flags (`-i`) to accomplish exactly that:
fw download "cni/testproject/subject1/session1" -i nifti -i bvec -i bval
This will generate an archive (.tar) file containing the requested hierarchy with only those files you explicitly need.
Example: Using quotes
Often times your source-path (that is the group/project/subject/session string the describes the location of your data in Flywheel) will have one or more spaces or special characters in it. To properly address that location using the CLI it's important to use quotes around the source path, like so:
fw download "cni/testproject/subject1/session1"
Example Download a single file with the CLI
So you have navigated to a container and your only desire is to download a single file from that container, the best way to do that is using the CLI with the 'files' spec filter.
Example: Download a single NIfTI file from an acquisition container:
fw download "test/Unsorted/s001/18591/T1w 1mm/files/18591_13_1.nii.gz"
The important bit here is the inclusion of "files" prior to the file name.
Using Sherlock and other Computers
We suggest that you use the Flywheel Command Line Interface (CLI) to transfer data to other compute resources, like Sherlock.
To download and install the CLI on Sherlock we use the `wget` tool, then `unzip` to extract the CLI resource package, and finally modify our `.bashrc` file to add the fw binary as an alias in our environment.
Step 1: Get the URL for the CLI package, which can be found on the "Profile" page within the Flywheel interface. To grab the URL, find the CLI section on the Profile page, right-click the Linux CLI Download link, and choose "Copy Link Address" (or similar). Once you have the download link address move to the next step.
Step 2: Log in to Sherlock and run the `wget` command, using the URL from Step 1 to download the CLI package.
mkdir -p flywheel/cli cd flywheel/cli wget //storage.googleapis.com/flywheel-dist/cli/<version>/fw-linux_amd64.zip
Step 3: Unpack the CLI archive and cleanup the downloaded package.
unzip fw-linux_amd64.zip mv linux_amd64/fw . rmdir linux_amd64 rm -f fw-linux_amd64.zip
Step 4: Modify your `.bashrc` file to add the fw CLI command to your environment, and source it to make the alias active. Note that this only needs to be done once.
echo -e "alias fw='$HOME/flywheel/cli/fw'" >> $HOME/.bashrc source $HOME/.bashrc
Step 5: Once the above steps are complete, you should be able to log in using the CLI and use it as described in the official documentation: //flywheelio.zendesk.com/hc/en-us/sections/360001596834-Command-Line-Interface. The best way to do this is navigate to your profile page in Flywheel, make sure that you have generated an API Key, and use the login command text that is provided for you there.
fw login <your API key>
Data Migration from NIMS
Before there was Flywheel, there was NIMS. Data have been preserved in NIMS and they can be migrated to Flywheel on an as-needed basis. Please inquire with Michael Perry for more information.
Support
Michael can help with most CNI Flywheel issues. If support is needed for Flywheel issues not related to the CNI, please email their help line: support@flywheel.io.