Spectro: Difference between revisions
imported>Laimab |
imported>Laimab |
||
| Line 32: | Line 32: | ||
The 13 topics for the experts' consensus recommendation papers include two general papers followed by articles on specific topics in advanced MRS grouped in three fields of research. | The 13 topics for the experts' consensus recommendation papers include two general papers followed by articles on specific topics in advanced MRS grouped in three fields of research. | ||
Two papers on the general background of methodology and publishing in MRS. | |||
Revision as of 01:30, 14 July 2021
MRS in Neurosciences: In-vivo spectroscopy methods and applications at CNI
Overview
Discoveries about the brain have implications for fields ranging from Business, Law, Psychology, and Education. The interest of measuring metabolic changes via MRS techniques and combining that information with functional MRI measurements continues to grow. CNI continues to support the research of its user community by providing state-of-the art data acquisition and state-of-the-art data management and analysis capabilities for in-vivo spectroscopy. Through collaborative efforts the special interest spectroscopy group at CNI has enabled education, participated in experimental design, and guided analyses and interpretation of results.
Spectroscopy
For users who are totally new to spectroscopy a good basic reference is MRI From Picture to Proton Donald W. McRobbie, Elizabeth A. Moore, Martin J. Graves, and Martin Prince, Cambridge University Press, Second Edition 2007, Chapter 15 (It's not just squiggles: in vivo spectroscopy). This chapter describes spectroscopy from the clinical side, but covers the basics for both data acquisition and processing.
Books:
In Vivo NMR Spectroscopy, 2nd Edition, Author: Robin de Graaf, Wiley Online Library
A recently published book Magnetic Resonance Spectroscopy - Tools for Neuroscience Research and Emerging Clinical Applications
An excellent spectroscopy resource website is https://mrshub.org/ The MRSHub is a curated collection of resources for the analysis of magnetic resonance spectroscopy data. It is maintained by the Committee for MRS Code and Data Sharing of the MR Spectroscopy Study Group of the International Society for Magnetic Resonance in Medicine (ISMRM).
Spectroscopy Literature
The most comprehensive collection of MRS papers have been published in 2021 in a special issue of NMR in Biomedicine - Advanced methodology for in vivo magnetic resonance spectroscopy - https://doi.org/10.1002/nbm.4504
Key points abstracted from the Editorial page of this special issue are:
"For this Special Issue, these 13 topics of advanced methodology in clinical and pre-clinical MRS have been reviewed by multi-institutional groups of authors, and a consensus overview of the most relevant facts and recommendations has been assembled for each of them. They now appear along with proffered review papers from single or smaller groups of authors and proffered research papers with a broad range of topics covering latest research results."
"The consensus articles that lay out the current state of the art and present recommendations for use in terms of several aspects of advanced MRS were written by teams of prime experts in these fields, where the teams had been instructed to self-organize under consideration of the width of the field and respecting geographic and gender aspects. However, limiting the number of authors to a size that allows productive interchange, it was not possible to include all potential expert authors as based on their publication records. Thus, it was decided for many of the papers to include a larger group of experts as a collaborator group to receive input from and to cross-check and support the experts' recommendations. None of these papers that include “experts' consensus recommendations” in their titles are to be considered as typical white papers where traditionally detailed advice on exact measurement protocols and parameters is given. This had not been our intention because the topics covered are of an advanced nature and hence inherently still touch many work-in-progress frontiers. Furthermore, all topics covered still present challenges in terms of identifying overall optimal methods as approved by the community as a whole. In contrast, the recommendations in these papers were arrived at as a consensus opinion among the specific group of experts without claiming validity for the whole community, and also with the possibility to vary the weight of advice from hints to more strongly encouraged recommendations."
"The experts' consensus papers are targeted primarily at two types of readers: on one hand, the non-MRS-specialized MR physicist or high-end user (eg a clinician or neuroscientist) commissioned with the task of implementing state-of-the-art MRS methods for a specific research or clinical target that go beyond the vendor-provided MRS techniques; on the other hand, MRS specialists who try to grasp the background and current advances in topics with which they are not intimately familiar and to provide starting points to them when they consider incorporating them in their research or technical developments."
The 13 topics for the experts' consensus recommendation papers include two general papers followed by articles on specific topics in advanced MRS grouped in three fields of research.
Two papers on the general background of methodology and publishing in MRS.
Recent Literature
Papers presented at the ISMRM MR Spectroscopy Study Group Virtual Meeting: NMR in Biomedicine Special Issue Experts' Recommendations: Where Consensus Was Reached and Where Dissent Prevailed, Part 1 (November 12, 2020)
Advanced methodology for in vivo magnetic resonance spectroscopy In‐Young Choi Roland Kreis https://doi.org/10.1002/nbm.4504 Editorial
B0 shimming for in vivo magnetic resonance spectroscopy: Experts' consensus recommendations Christoph Juchem, Cristina Cudalbu, Robin A. de Graaf, Rolf Gruetter, Anke Henning, Hoby P. Hetherington, Vincent O. Boer NMR in Biomedicine 2020;e4350 https://doi.org/10.1002/nbm.4350
Advanced magnetic resonance spectroscopic neuroimaging: Experts' consensus recommendations Andrew A. Maudsley, Ovidiu C. Andronesi, Peter B. Barker, Alberto Bizzi, Wolfgang Bogner, Anke Henning, Sarah J. Nelson, Stefan Posse, Dikoma C. Shungu, Brian J. Soher NMR in Biomedicine 2020;e4309 https://doi.org/10.1002/nbm.4309
Motion correction methods for MRS: experts' consensus recommendations Ovidiu C. Andronesi, Pallab K. Bhattacharyya, Wolfgang Bogner, In‐Young Choi, Aaron T. Hess, Phil Lee, Ernesta M. Meintjes, M. Dylan Tisdall, Maxim Zaitzev, André van der Kouwe NMR in Biomedicine 2020;e4364 https://doi.org/10.1002/nbm.4364
Preprocessing, analysis and quantification in single‐voxel magnetic resonance spectroscopy: experts' consensus recommendations Jamie Near Ashley D. Harris Christoph Juchem Roland Kreis Małgorzata Marjańska Gülin Öz Johannes Slotboom Martin Wilson Charles Gasparovic NMR in Biomedicine 2020;e4257 https://doi.org/10.1002/nbm.4257
Proton magnetic resonance spectroscopy in skeletal muscle: Experts' consensus recommendations Martin Krššák, Lucas Lindeboom, Vera Schrauwen‐Hinderling, Lidia S. Szczepaniak, Wim Derave, Jesper Lundbom, Douglas Befroy, Fritz Schick, Jürgen Machann, Roland Kreis, Chris Boesch NMR in Biomedicine 2020;e4266 https://doi.org/10.1002/nbm.4266
31P magnetic resonance spectroscopy in skeletal muscle: Experts' consensus recommendations Martin Meyerspeer, Chris Boesch, Donnie Cameron, Monika Dezortová, Sean C. Forbes, Arend Heerschap, Jeroen A.L. Jeneson, Hermien E. Kan, Jane Kent, Gwenaël Layec, Jeanine J. Prompers, Harmen Reyngoudt, Alison Sleigh, Ladislav Valkovič, Graham J. Kemp, Experts' Working Group on 31P MR Spectroscopy of Skeletal Muscle NMR in Biomedicine 2020;e4246 https://doi.org/10.1002/nbm.4246
Other review papers in the special issue now online
Terminology and concepts for the characterization of in vivo MR spectroscopy methods and MR spectra: Background and experts' consensus recommendations Roland Kreis, Vincent Boer, In‐Young Choi, Cristina Cudalbu, Robin A. de Graaf, Charles Gasparovic,,Arend Heerschap, Martin Krššák, Bernard Lanz, Andrew A. Maudsley, Martin Meyerspeer, Jamie Near, Gülin Öz, Stefan Posse, Johannes Slotboom, Melissa Terpstra, Ivan Tkáč, Martin Wilson, Wolfgang Bogner, Experts' Working Group on Terminology for MR Spectroscopy NMR in Biomedicine 2020;e4347 https://doi.org/10.1002/nbm.4347
Spectral editing in 1H magnetic resonance spectroscopy: Experts' consensus recommendations In‐Young Choi, Ovidiu C. Andronesi, Peter Barker, Wolfgang Bogner, Richard A. E. Edden, Lana G. Kaiser, Phil Lee, Małgorzata Marjańska, Melissa Terpstra, Robin A. de Graaf NMR in Biomedicine 2020;e4411 https://doi.org/10.1002/nbm.4411
Magnetic resonance spectroscopy in the rodent brain: Experts' consensus recommendations Bernard Lanz, Alireza Abaei, Olivier Braissant, In‐Young Choi, Cristina Cudalbu, Pierre‐Gilles Henry, Rolf Gruetter, Firat Kara, Kejal Kantarci, Phil Lee, Norbert W. Lutz, Małgorzata Marjańska, Vladimír Mlynárik, Volker Rasche, Lijing Xin, Julien Valette, the Experts' Working Group on Magnetic resonance spectroscopy in the rodent brain NMR in Biomedicine 2020;e4325 https://doi.org/10.1002/nbm.4325
Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: Experts' consensus recommendations Gülin Öz Dinesh K. Deelchand Jannie P. Wijnen Vladimír Mlynárik Lijing Xin Ralf Mekle Ralph Noeske Tom W.J. Scheenen Ivan Tkáč the Experts' Working Group on Advanced Single Voxel 1H MRS https://doi.org/10.1002/nbm.4236 NMR in Biomedicine. 2020;e4236
Data Acquisition and Processing Tools
Spectroscopy Sequences
The Stanford CNI effort has become a best practice through a community effort with spectroscopy expertise from CNI staff, MRI scientists, and an expanding user community. Current spectroscopy sequences include methods both for edited GABA (gamma-Aminobutyric acid) specific data acquisition (MEGA-PRESS [1] and IM-SPECIAL [3]), and for multi-metabolite data acquisition (Optimized-PRESS [4], [5], [6]). Additional sequences such as semi-LASER [9], [10], [11], [12], [13] are being evaluated and added as newer data acquisition methods.
| Spectroscopy Sequence | Measured Metabolites | Analysis Methods |
|---|---|---|
| MEGA-PRESS [1] | GABA+, Glx(Glutamate, Glutmine) | Gannet [2] |
| IM-SPECIAL [3] | GABA, Glu(Glutamate), Glx(Glutamate, Glutamine) | Sequence specific Matlab code |
| Optimized-PRESS [4], [5], [6] | All metabolites | Sequence specific Matlab code, LCModel fitting [7] |
| semi-LASER [9], [10], [11], [12], [13] | All metabolites | Sequence specific Matlab code, LCModel fitting [7] |
References
[1] Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R (1998) Simultaneous in vivo spectral editing and water suppression, NMR Biomed. 11:266–272 https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291099-1492%28199810%2911%3A6%3C266%3A%3AAID-NBM530%3E3.0.CO%3B2-J
[2] Richard A.E. Edden, Nicolaas A.J. Puts, Ashley D. Harris, Peter B. Barker, and C. John Evans (2014) Gannet: A Batch-Processing Tool for the Quantitative Analysis of Gamma-Aminobutyric Acid–Edited MR Spectroscopy Spectra, Journal of Magnetic Resonance Imaging 40:1445–1452 https://doi.org/10.1002/jmri.24478
[3] Gu M, Hurd R, Noeske R, Baltusis L, Hancock R, Sacchet MD, Gotlib IH, Chin FT, Spielman DM (2018) GABA editing with macromolecule suppression using an improved MEGA-SPECIAL sequence, Magnetic Resonance in Medicine 79:41-47 https://doi.org/10.1002/mrm.26691
[4] Webb PG, Sailasuta N, Kohler SJ, Raidy T, Moats RA, Hurd R (1994) Automated single voxel proton MRS: technical development and multisite verification, Magnetic Resonance in Medicine 31(4):365-373 https://doi.org/10.1002/mrm.1910310404
[5] Bodenhausen G, Freeman R, Turner DL (1977) Suppression of artifacts in two dimensional J spectroscopy, Journal of Magnetic Resonance Imaging 27:511-514 https://doi.org/10.1016/0022-2364(77)90016-6
[6] Tran TK, Vigneron DB, Sailasuta N, Tropp J, Le Roux P, Kurhanewicz J, Nelson S, Hurd R (2000) Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer, Magnetic Resonance in Medicine 43(1):23-33. https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291522-2594%28200001%2943%3A1%3C23%3A%3AAID-MRM4%3E3.0.CO%3B2-E
[7] Provencher SW (2001) Automatic quantitation of localized in vivo1H spectra with LCModel, NMR in Biomedicine 14(4):260-264 https://doi.org/10.1002/nbm.698
[8] Young Woo Park, Dinesh K. Deelchand, James M. Joers, Brian Hanna, Adam Berrington, Joseph S. Gillen, Kejal Kantarci, Brian J. Soher, Peter B. Barker, HyunWook Park, Gulin Oz, Christophe Lenglet (2018) AutoVOI: real-time automatic prescription of volume-of-interest for single voxel spectroscopy, Magn. Reson. Med. 80:1787–1798 https://doi.org/10.1002/mrm.27203
[9] Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A. (2018) Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses, Magn. Reson. Med. 59(1):1-6 https://doi.org/10.1002/mrm.21302
[10] Oz G, Tkac I. (2011) Short-echo, single-shot, full-intensity proton magnetic resonance spectroscopy for neurochemical profiling at 4 T: validation in the cerebellum and brainstem, Magn. Reson. Med. 65(4):901-910 https://doi.org/10.1002/mrm.22708
[11] Martin Wilson, Ovidiu Andronesi, Peter B. Barker, Robert Bartha, Alberto Bizzi, Patrick J. Bolan, Kevin M. Brindle, In-Young Choi, Cristina Cudalbu, Ulrike Dydak, Uzay E. Emir, Ramon G. Gonzalez, Stephan Gruber, Rolf Gruetter, Rakesh K. Gupta, Arend Heerschap, Anke Henning,Hoby P. Hetherington, Petra S. Huppi, Ralph E. Hurd, Kejal Kantarci, Risto A Kauppinen, Dennis W. J. Klomp, Roland Kreis, Marijn J. Kruiskamp, Martin O. Leach, Alexander P. Lin, Peter R. Luijten, Malgorzata Marjanska, Andrew A. Maudsley, Dieter J. Meyerhoff, Carolyn E. Mountford, Paul G. Mullins, James B. Murdoch, Sarah J. Nelson, Ralph Noeske, Gulin Oz, Julie W. Pan, Andrew C. Peet, Harish Poptani, Stefan Posse, Eva-Maria Ratai, Nouha Salibi, Tom W. J. Scheenen, Ian C. P. Smith, Brian J. Soher, Ivan Tkac, Daniel B. Vigneron, Franklyn A. Howe (2019) Methodological consensus on clinical proton MRS of the brain:Review and recommendations, Magn. Reson. Med. 82:527–550 https://doi.org/10.1002/mrm.27742
[12] Dinesh K. Deelchand, Adam Berrington, Ralph Noeske, James M. Joers, Arvin Arani, Joseph Gillen, Michael Schar, Jon-Fredrik Nielsen, Scott Peltier, Navid Seraji-Bozorgzad, Karl Landheer, Christoph Juchem, Brian Soher, Douglas C. Noll, Kejal Kantarci, Eva M. Ratai, Thomas H. Marecii, Peter B. Barker, Gulin Oz (2019) Across-vendor standardization of semi-LASER for single-voxel MRS at 3T, NMR in Biomedicine. e4218 https://doi.org/10.1002/nbm.4218
[13] Gulin Oz, Dinesh K. Deelchand, Jannie P. Wijnen, Vladimir Mlynarik, Lijing Xin, Ralf Mekle, Ralph Noeske, Tom W.J. Scheenen, Ivan Tkac, the Experts' Working Group on Advanced Single Voxel 1H MRS (2020) Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: Experts' consensus recommendations, NMR in Biomedicine. e4236 https://doi.org/10.1002/nbm.4236
[14] L. Ryner, J Sorenson, M.A. Thomas (1995) Localized 2D J-resolved H-1 MR spectroscopy: Strong coupling effects in vitro and in vivo, Magn. Reson. Imaging 13:853–869 https://doi.org/10.1016/0730-725X(95)00031-B
[15] R. Schulte, T. Lange, J. Beck, D. Meier, P. Boesiger (2006) Improved two-dimensional J-resolved spectroscopy, NMR Biomed. 19:264–270 https://doi.org/10.1002/nbm.1027
[16] Krish Krishnamurthy (2013) CRAFT (complete reduction to amplitude frequency table) – robust and time efficient Bayesian approach for quantitative mixture analysis by NMR, Magn. Reson. Chem. 51: 821–829 https://doi.org/10.1002/mrc.4022
Data Management
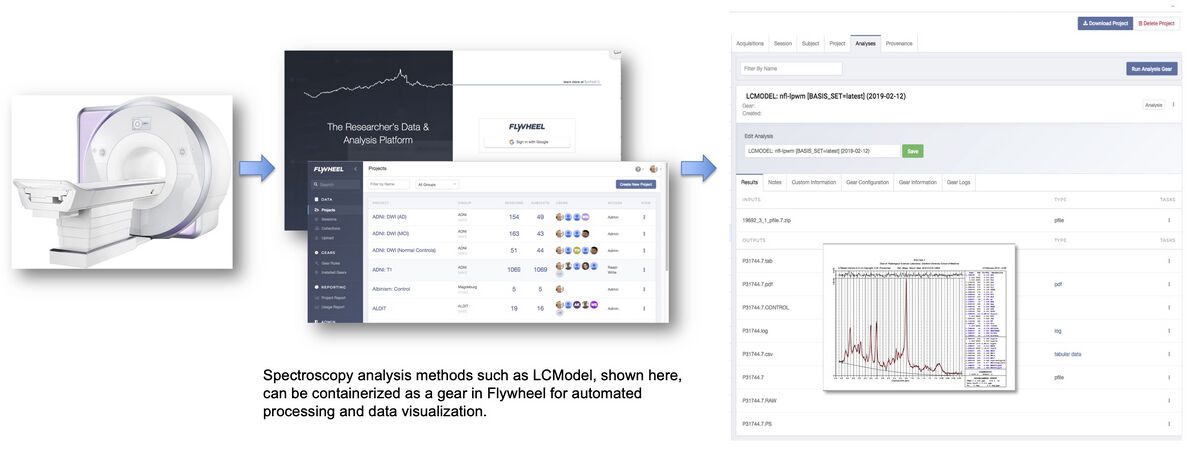
CNI currently uses Flywheel as its data base management system. A critical feature of this data base management is the ability to share computational methods within the system. The CNI now provides a combination of data repository and integrated open source processing tools such as Gannet [2] and LCModel [7]. This combination of tools supports scientific transparency for both data and computational sharing. Spectroscopy analysis methods such as LCModel can be containerized as a gear in Flywheel for automated processing and data visualization.
Collecting Data - Sequences and protocols
Current spectroscopy sequences include methods for multi-metabolite data acquisition (Optimized-PRESS [4], [5], [6]) and semi-LASER [9], [10], [11], [12], [13]) and for edited GABA (gamma-Aminobutyric acid) specific data acquisition (MEGA-PRESS [1])
This protocol (spectro-protocol-1 located in the CNI Other tab on the scanner) contains the optimized-PRESS and sLaser sequences for data collection for all metabolites
optimized-PRESS
[4] Webb PG, Sailasuta N, Kohler SJ, Raidy T, Moats RA, Hurd R (1994) Automated single voxel proton MRS: technical development and multisite verification, Magnetic Resonance in Medicine 31(4):365-373 https://doi.org/10.1002/mrm.1910310404
[5] Bodenhausen G, Freeman R, Turner DL (1977) Suppression of artifacts in two dimensional J spectroscopy, Journal of Magnetic Resonance Imaging 27:511-514 https://doi.org/10.1016/0022-2364(77)90016-6
[6] Tran TK, Vigneron DB, Sailasuta N, Tropp J, Le Roux P, Kurhanewicz J, Nelson S, Hurd R (2000) Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer, Magnetic Resonance in Medicine 43(1):23-33. https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291522-2594%28200001%2943%3A1%3C23%3A%3AAID-MRM4%3E3.0.CO%3B2-E
 |
 |
 |
s-Laser
High field (3T) and ultra-high field (7T) are ideal field strength for MR spectroscopy due to the higher spectral resolution and higher signal that can be achieved. But with these advantages comes a higher B1-inhomogeneity and larger Chemical Shift Displacement Error (CSDE). Semi-LASER is a double spin-echo MRS technique like the established PRESS (GE product name Probe-P) technique that uses a slice selective non-adiabatic excitation and two pairs of adiabatic slice selective refocusing pulses for volume selection. The adiabatic behavior of the RF pulses addresses the B1-inhomogeneity problem while the increased bandwidth of these pulses reduces the CSDE.
[9] Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A. (2018) Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses, Magn. Reson. Med. 59(1):1-6 https://doi.org/10.1002/mrm.21302
[10] Oz G, Tkac I. (2011) Short-echo, single-shot, full-intensity proton magnetic resonance spectroscopy for neurochemical profiling at 4 T: validation in the cerebellum and brainstem, Magn. Reson. Med. 65(4):901-910 https://doi.org/10.1002/mrm.22708
[11] Martin Wilson, Ovidiu Andronesi, Peter B. Barker, Robert Bartha, Alberto Bizzi, Patrick J. Bolan, Kevin M. Brindle, In-Young Choi, Cristina Cudalbu, Ulrike Dydak, Uzay E. Emir, Ramon G. Gonzalez, Stephan Gruber, Rolf Gruetter, Rakesh K. Gupta, Arend Heerschap, Anke Henning,Hoby P. Hetherington, Petra S. Huppi, Ralph E. Hurd, Kejal Kantarci, Risto A Kauppinen, Dennis W. J. Klomp, Roland Kreis, Marijn J. Kruiskamp, Martin O. Leach, Alexander P. Lin, Peter R. Luijten, Malgorzata Marjanska, Andrew A. Maudsley, Dieter J. Meyerhoff, Carolyn E. Mountford, Paul G. Mullins, James B. Murdoch, Sarah J. Nelson, Ralph Noeske, Gulin Oz, Julie W. Pan, Andrew C. Peet, Harish Poptani, Stefan Posse, Eva-Maria Ratai, Nouha Salibi, Tom W. J. Scheenen, Ian C. P. Smith, Brian J. Soher, Ivan Tkac, Daniel B. Vigneron, Franklyn A. Howe (2019) Methodological consensus on clinical proton MRS of the brain:Review and recommendations, Magn. Reson. Med. 82:527–550 https://doi.org/10.1002/mrm.27742
[12] Dinesh K. Deelchand, Adam Berrington, Ralph Noeske, James M. Joers, Arvin Arani, Joseph Gillen, Michael Schar, Jon-Fredrik Nielsen, Scott Peltier, Navid Seraji-Bozorgzad, Karl Landheer, Christoph Juchem, Brian Soher, Douglas C. Noll, Kejal Kantarci, Eva M. Ratai, Thomas H. Marecii, Peter B. Barker, Gulin Oz (2019) Across-vendor standardization of semi-LASER for single-voxel MRS at 3T, NMR in Biomedicine. e4218 https://doi.org/10.1002/nbm.4218
[13] Gulin Oz, Dinesh K. Deelchand, Jannie P. Wijnen, Vladimir Mlynarik, Lijing Xin, Ralf Mekle, Ralph Noeske, Tom W.J. Scheenen, Ivan Tkac, the Experts' Working Group on Advanced Single Voxel 1H MRS (2020) Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: Experts' consensus recommendations, NMR in Biomedicine. e4236 https://doi.org/10.1002/nbm.4236
 |
 |
 |
This protocol (spectro-protocol-editing-1 located in the CNI Other tab on the scanner) contains the MEGA-PRESS sequence for data collection for GABA edited data

MEGA-PRESS
[1] Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R (1998) Simultaneous in vivo spectral editing and water suppression, NMR Biomed. 11:266–272 https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291099-1492%28199810%2911%3A6%3C266%3A%3AAID-NBM530%3E3.0.CO%3B2-J
 |
 |
 |
 |
 |
Processing and Data Analyis with Flywheel
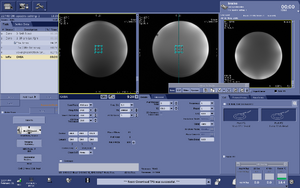
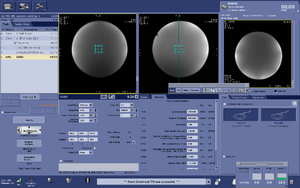
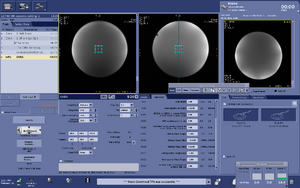
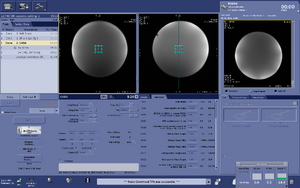
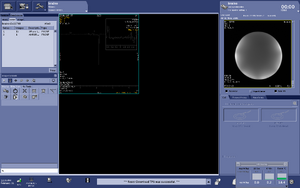
Note: For users using the CNI Optimized PRESS "nfl" sequence with LCModel data analysis - Users who are in groups that are not themselves Flywheel "Lab" customers will need to download their results from the session tab now (not the analyses tab as was done before and shown in the figure below)
If you are using Google Chrome as the browser to download the processed data, then you will need to add the extension .zip to the downloaded file name. This operation does not not need to be done if using Safari or Firefox browsers.
MEGA-PRESS results will continue to be in the analyses tab.

Processing and Analyzing GABA Data - non-Flywheel methods
(1) Analysis of GABA Data collected with the MEGA-PRESS sequence with Gannet
To analyze the data from MEGA-PRESS experiments, we recommend Gannet, a batch-analysis tool for GABA-edited MRS data. The Gannet code and instruction manual can be downloaded from here gabamrs.blogspot.com Additional and newer information can be found here www.gabamrs.com and here www.gabamrs.com/about/ Note that Matlab with Optimization and Statistics Toolboxes must be installed on your computer prior to downloading the Gannet2.0 code.
Note that following change needs to be made to the Gannet code in order to correctly analyze data obtained the CNI MEGA-PRESS sequence gabamrs.blogspot.com (When Gannet goes wrong section)
From gabamrs blogspot -
Solution 2: ON and OFF are incorrectly identified.
If the creatine stripe is correct (red on blue; spectra phased positively) and the difference spectra are still negative, then the issue is the ordering of ON and OFF spectra. The simple solution is to change the MRS_struct.p.onofforder parameter in GannetPreInitialise.m. It is either 'onfirst' or 'offfirst' depending on the acquisition order.
So here's the summary:
"Gannet makes my GABA difference spectra negative".
"Are your creatine signals phased positively?".
If yes, change MRS_struct.p.onofforder; if no, change MRS_struct.p.WaterPositive.
Notes on Spectro Scans on DV26
Prescribing a rotated voxel
At the moment GE’s DV26 software platform release doesn’t support voxel rotation for spectroscopy sequences. A workaround however is available:
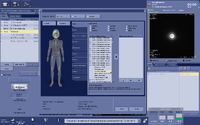
(1) After the 3-plane reconstruction of the T1w images, setup a 3D imaging scan. We saved a template of this 3D scan in the protocol “CNI Example Spectroscopy” and named it as “Voxel prescription”. Add this sequence to your protocol and put it before the MRS scan.
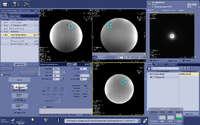
(2) Setup the voxel prescription scan. The Scan Plane is set to “Oblique”, FOV and Locs per Slab are set as close to the voxel size of the spectro scan as possible. Prescribe the 3D box, place it to the desired location and rotate to the right angle. Because the box is usually bigger than the voxel size you want to prescribe, the coverage you see in this step is not accurate. It’s crucial to get the correct orientation of the box. Save Rx.
(3) Setup the MRS scan. Copy Rx from the voxel prescription scan. By default the Mode Filter in the Copy Rx is set to “MRS”, you need to change it to “All” or “3D” in order to see the voxel prescription scan in the Copy Rx list. Select it and accept.
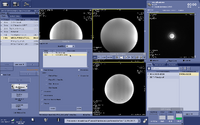
(4) Adjust voxel location and size. You can move the box around and change its size on the graphic interface, or set its coordinates and size by setting the Center and Length in X, Y, Z. If you want to adjust the orientation of the voxel, you need to go back to the 3D voxel prescription scan, and repeat step 2-4 again. After you finish setting up the MRS scan, Save Rx and proceed with scanning.

Extracting voxel prescription information retrospectively
The prescription information of the voxel of an MRS scan is saved in the header of the raw data (p-file). Specifically, the x, y, z dimensions of the voxel are stored in header fields rdb_hdr_image.user8, rdb_hdr_image.user9, rdb_hdr_image.user10; the x, y, z locations of the center of the voxel are stored in header fields rdb_hdr_image.user11, rdb_hdr_image.user12, rdb_hdr_image.user13, in RAS coordinates (e.g. if the voxel is at L26.0 then rdb_hdr_image.user11 will record -26.0). The unit is mm.
To look for this information on Flywheel, open the information window of the p-file, and search for op_user_8, op_user_9, op_user_10, etc.