Spectro: Difference between revisions
imported>Laimab |
imported>Laimab |
||
| Line 321: | Line 321: | ||
|} | |} | ||
''' Screen shots of parameters for optimized-press sequence''' | |||
{| | {| | ||
Revision as of 00:52, 13 August 2021
MRS in Neurosciences: In-vivo spectroscopy methods and applications at CNI
Overview
Discoveries about the brain have implications for fields ranging from Business, Law, Psychology, and Education. The interest of measuring metabolic changes via MRS techniques and combining that information with functional MRI measurements continues to grow. CNI continues to support the research of its user community by providing state-of-the art data acquisition and state-of-the-art data management and analysis capabilities for in-vivo spectroscopy. Through collaborative efforts the special interest spectroscopy group at CNI has enabled education, participated in experimental design, and guided analyses and interpretation of results.
Examples of studies at CNI using in-vivo spectroscopy techniques include characterization of biomarkers following transcranial magnetic stimulation, and metabolite characterization for conditions such as substance addiction, pain, depression, and various forms of dementia. These studies illustrate integrated data acquisition and data processing tools accessible to the CNI user community which simplify the use and sharing of spectroscopy results in neuroimaging applications.
Association between anterior cingulate neurochemical concentration and individual differences in hypnotizability DeSouza DD, Stimpson K, Baltusis L, Sacchet MD, Gu M, Hurd R, Wu H, Yeomans DC, Williams N, Spiegel D. Association between anterior cingulate neurochemical concentration and individual differences in hypnotizability. Cerebral Cortex, Volume 30, Issue 6, June 2020, Pages 3644–3654, https://doi.org/10.1093/cercor/bhz332
The CNI continues to support the research of its user community by developing and incorporating for general use new data acquisition and data analysis capabilities.
GABA editing with macromolecule suppression using an improved MEGA-SPECIAL sequence Gu M, Hurd R, Noeske R, Baltusis L, Hancock R, Sacchet MD, Gotlib IH, Chin FT, Spielman DM (2018) GABA editing with macromolecule suppression using an improved MEGA-SPECIAL sequence, Magnetic Resonance in Medicine 79:41-47 https://doi.org/10.1002/mrm.26691
Spectroscopy Websites and Literature
Websites:
An excellent spectroscopy resource website is https://mrshub.org/ The MRSHub is a curated collection of resources for the analysis of magnetic resonance spectroscopy data. It is maintained by the Committee for MRS Code and Data Sharing of the MR Spectroscopy Study Group of the International Society for Magnetic Resonance in Medicine (ISMRM).
To analyze the data from MEGA-PRESS experiments Gannet can be used. It is a batch-analysis tool for GABA-edited MRS data. The Gannet code and instruction manual can be downloaded from here gabamrs.blogspot.com Additional information can be found here www.gabamrs.com and here www.gabamrs.com/about/ Note that Matlab with Optimization and Statistics Toolboxes must be installed on your computer prior to downloading the Gannet2.0 code.
Books:
In Vivo NMR Spectroscopy, 2nd Edition, Author: Robin de Graaf, Wiley Online Library
The recently updated 3rd edition: In Vivo NMR Spectroscopy, 3rd Edition, Author: Robin de Graaf, Wiley Online Library
The 2014 book Magnetic Resonance Spectroscopy - Tools for Neuroscience Research and Emerging Clinical Applications
Papers:
A very comprehensive collection of MRS papers have been published in 2021 in a special issue of NMR in Biomedicine - Advanced methodology for in vivo magnetic resonance spectroscopy - https://doi.org/10.1002/nbm.4504
Key points abstracted from the Editorial page of this special issue are summarized here and the most relevant papers to the CNI spectroscopy tools are in bold type:
"For this Special Issue, these 13 topics of advanced methodology in clinical and pre-clinical MRS have been reviewed by multi-institutional groups of authors, and a consensus overview of the most relevant facts and recommendations has been assembled for each of them. They now appear along with proffered review papers from single or smaller groups of authors and proffered research papers with a broad range of topics covering latest research results."
"The consensus articles that lay out the current state of the art and present recommendations for use in terms of several aspects of advanced MRS were written by teams of prime experts in these fields, where the teams had been instructed to self-organize under consideration of the width of the field and respecting geographic and gender aspects. However, limiting the number of authors to a size that allows productive interchange, it was not possible to include all potential expert authors as based on their publication records. Thus, it was decided for many of the papers to include a larger group of experts as a collaborator group to receive input from and to cross-check and support the experts' recommendations. None of these papers that include “experts' consensus recommendations” in their titles are to be considered as typical white papers where traditionally detailed advice on exact measurement protocols and parameters is given. This had not been our intention because the topics covered are of an advanced nature and hence inherently still touch many work-in-progress frontiers. Furthermore, all topics covered still present challenges in terms of identifying overall optimal methods as approved by the community as a whole. In contrast, the recommendations in these papers were arrived at as a consensus opinion among the specific group of experts without claiming validity for the whole community, and also with the possibility to vary the weight of advice from hints to more strongly encouraged recommendations."
"The experts' consensus papers are targeted primarily at two types of readers: on one hand, the non-MRS-specialized MR physicist or high-end user (eg a clinician or neuroscientist) commissioned with the task of implementing state-of-the-art MRS methods for a specific research or clinical target that go beyond the vendor-provided MRS techniques; on the other hand, MRS specialists who try to grasp the background and current advances in topics with which they are not intimately familiar and to provide starting points to them when they consider incorporating them in their research or technical developments."
The 13 topics for the experts' consensus recommendation papers include two general papers followed by articles on specific topics in advanced MRS grouped in three fields of research.
*Two papers on the general background of methodology and publishing in MRS.
**Terminology and concepts in MRS Kreis R, Boer V, Choi IY, et al. Terminology and concepts for the characterization of in vivo MR spectroscopy methods and MR spectra: background and experts' consensus recommendations. NMR Biomed. 2020;e4347. https://doi.org/10.1002/nbm.4347 The first article of the special issue has the broad scope of presenting the basic framework of concepts and terminology in the field of in vivo MRS, which will hopefully help to standardize the language used and thus prevent misunderstandings in future publications in the field. It also contains suggestions for the use of abbreviations and provides references to the origin of concepts, methods and common abbreviations that at times appear without reference.
**Reporting standards Lin A, Andronesi O, Bogner W, et al. Minimum reporting standards for in vivo magnetic resonance spectroscopy (MRSinMRS): experts' consensus recommendations. NMR Biomed. 2021;e4484. https://doi.org/10.1002/nbm.4484 The second paper of a more general scope is concerned with minimal reporting guidelines for papers that include MRS data, whether published in MR literature or even more so in clinical journals. It has been a common experience for many of the experienced MRS methods experts to find essential details about employed MRS methods missing from published papers or in manuscripts submitted for review. Hence, Lin et al have assembled a list of items that the authors, who are all experienced referees, suggest should be covered in the methods section of future papers including MRS results. For ease of use, the paper also contains a checklist that could be submitted along with a manuscript as supplemental material or as a way to check that all vital information is provided in the methods section.
Checklist from https://doi.org/10.1002/nbm.4484
MRI system description
a. Field strength, eg 1.5 T, 3 T, 7 T, 9.4 T
b. Manufacturer, eg General Electric, Philips, Siemens, Toshiba
c. Model, eg General Electric Signa HD/X/T/de, Optima MR450/MR450W, Discovery MR750/MR750W, Signa Premier; Siemens Biograph mMR,Magnetom, Aera, Espree, Prisma, Skyra, Trio, Verio, Magnetom 7 T, Terra; Phillips Ingenia 1.5 T S, 3 T X/S, Elition 3 T X/S, Ambition 1.5 T X/S, Achieva 1.5 T/3 T. Software version, eg Siemens VB17A, VD19, VE11C; General Electric 12x-24x; Phillips Release 5, 5.1 (R1-3), 5.6, 6
d. RF coils used (nuclei, number of channels, type, body part), eg 1H, 31P, 13C, 31P-1H; type, eg head/neck, torso, knee; if not manufacturer, design, eg butterfly, quadrature etc
e. Additional hardware, eg shim inserts, dielectric pads
Acquisition parameters in full
i. Pulse sequence, eg spin-echo, point resolved spectroscopy (PRESS), stimulated echo acquisition mode (STEAM), semi-LASER, etc.
ii. Location of volume(s) of interest (VOI(s)), eg posterior cingulate gyrus, M. tibialis anterior, internal capsule of prostate, etc. A figure that displays the VOI on anatomic images is recommended.
iii. Nominal VOI size [cm3, mm3], eg 40 × 40 × 10 mm3.
iv. Repetition time (TR), echo time (TE) [ms, s]; if STEAM, mixing time (TM).
v. Total number of excitations per spectrum.
vi. Additional sequence parameters, eg
i. Spectral width [Hz, kHz] and number of data points
ii. Frequency offset (if any)
iii. If magnetic resonance spectroscopic imaging (MRSI): specification of two-dimensional (2D) or three-dimensional (3D) spatial mapping, field of view (FOV), matrix size, acceleration factor, sampling/reconstruction method (eg parallel imaging, compressed sensing, spatial spectral encoding, etc), nominal and effective (ie final) voxel volumes, flip angles for fast MRSI
iv. For multidimensional acquisitions, number of encodings in the second spectral dimension
v. For editing methods, editing pulse information including pulse shape, bandwidth and offset frequency
vi. For multinuclear sequences: details of decoupling or polarization transfer sequences and related parameters.
f. Water suppression method (and any other suppression methods used, eg lipid suppression, outer volume suppression).
g. Shimming method, reference peak used for assessing shim performance, and thresholds for “acceptance of shim” chosen.
h. Triggering method, if used (respiratory, peripheral, cardiac triggering, including device used and delays).
i. Frequency and motion correction methods, if used (prospective or retrospective, external tracker or navigator method).
Spectral quantification methods and parameters
a. Software package used to reconstruct and analyze the MRS data including MR manufacture software (eg General Electric PROBE, Siemens Syngo, or Phillips SpectroView) and/or third-party software packages (eg LCModel, jMRUI, TARQUIN, SIVIC, INSPECTOR, FID-A, BrainSpec, MIDAS, GANNET)
b. Deviations in processing steps from quoted reference or product defaults
c. Quantitative output measures
d. Quantification references and assumptions, model fitting assumptions
Quality assurance. Studies must include the following
a. Reported variables (SNR, linewidth, and description of how they were obtained)
b. Data exclusion criteria
c. Other quality measures from fitting software are also recommended (eg standard deviation (SD), Cramér-Rao lower bound (CRLB)), and/or the robustness of the measures gained (repeatability measures if known)
d. Figure showing representative spectra
*Four papers dealing with commonly used MRS acquisition sequences and processing methods.
**Advanced single-voxel MRS Oz G, Deelchand DK, Wijnen JP, et al. Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: experts' consensus recommendations. NMR Biomed. 2020;e4236. https://doi.org/10.1002/nbm.4236
**Advanced neuro-MRSI Maudsley AA, Andronesi OC, Barker PB, et al. Advanced magnetic resonance spectroscopic neuroimaging: experts' consensus recommendations. NMR Biomed. 2020;e4309. https://doi.org/10.1002/nbm.4309
**Spectral editing Choi IY, Andronesi OC, Barker P, et al. Spectral editing in 1H magnetic resonance spectroscopy: experts' consensus recommendations. NMR Biomed. 2020;e4411. https://doi.org/10.1002/nbm.4411
**Processing and quantification Near J, Harris AD, Juchem C, et al. Preprocessing, analysis and quantification in single-voxel magnetic resonance spectroscopy: experts' consensus recommendations. NMR Biomed. 2020;e4257. https://doi.org/10.1002/nbm.4257
- Three papers related to MRS techniques that are critical to ensure the quality of data.
- Water and lipid suppression Tkac I, Deelchand D, Dreher W, et al. Water and lipid suppression techniques for advanced 1H MRS and MRSI of the human brain: experts' consensus recommendations. NMR Biomed. 2020;e4459. https://doi.org/10.1002/nbm.4459
- B0 shimming Juchem C, Cudalbu C, de Graaf RA, et al. B0 shimming for in vivo magnetic resonance spectroscopy: experts' consensus recommendations. NMR Biomed. 2020;e4350. https://doi.org/10.1002/nbm.4350
- Motion correction Andronesi OC, Bhattacharyya PK, Bogner W, et al. Motion correction methods for MRS: experts' consensus recommendations. NMR Biomed. 2020;e4364. https://doi.org/10.1002/nbm.4364
- Four papers appertaining to special targets of MRS studies.
- Macromolecular signals and ways to accommodate them in1H MR brain spectra Cudalbu C, Behar KL, Bhattacharyya PK, et al. Contribution of macromolecules to brain 1H MR spectra: experts' consensus recommendations. NMR Biomed. 2020;e4393. https://doi.org/10.1002/nbm.4393
- 1H MRS of skeletal muscle Krssak M, Lindeboom L, Schrauwen-Hinderling V, et al. Proton magnetic resonance spectroscopy in skeletal muscle: experts' consensus recommendations. NMR Biomed. 2020;e4266. https://doi.org/10.1002/nbm.4266
- 31P MRS of skeletal muscle Meyerspeer M, Boesch C, Cameron D, et al. 31P magnetic resonance spectroscopy in skeletal muscle: experts' consensus recommendations. NMR Biomed. 2020;e4246. https://doi.org/10.1002/nbm.4246
- Specifics for MRS methods in preclinical applications Lanz B, Abaei A, Braissant O, et al. Magnetic resonance spectroscopy in the rodent brain: experts' consensus recommendations. NMR Biomed. 2020;e4325. https://doi.org/10.1002/nbm.4325
- This special issue also includes authoritative reviews of the current state of
- fast MRSI methodology Bogner W, Otazo R, Henning A. Accelerated MR spectroscopic imaging—a review of current and emerging techniques. NMR Biomed. 2020;e4314. https://doi.org/10.1002/nbm.4314
- hyperpolarized 13C MRI and MRS Crane JC, Gordon JW, Chen H-Y, et al. Hyperpolarized 13C MRI data acquisition and analysis in prostate and brain at University of California, San Francisco. NMR Biomed. 2020;e4280. https://doi.org/10.1002/nbm.4280
- functional MRS in rodents Just N. Proton functional magnetic resonance spectroscopy in rodents. NMR Biomed. 2020;e4254. https://doi.org/10.1002/nbm.4254
- Fifteen proferred original research contributions complete this special issue. They cover latest research results in terms of novel advanced MRS methodology or its application. The methodology addresses issues in
**standardization - Deelchand DK, Berrington A, Noeske R, et al. Across-vendor standardization of semi-LASER for single-voxel MRS at 3T. NMR Biomed. 2019;e4218. https://doi.org/10.1002/nbm.4218
- diffusion MRS Lundell H, Ingo C, Dyrby TB, Ronen I. Cytosolic diffusivity and microscopic anisotropy of N-acetyl aspartate in human white matter with diffusion-weighted MRS at 7 T. NMR Biomed. 2020;e4304. https://doi.org/10.1002/nbm.4304 Hanstock C, Beaulieu C. Rapid acquisition diffusion MR spectroscopy of metabolites in human brain. NMR Biomed. 2020;e4270. https://doi.org/10.1002/nbm.4270 Genovese G, Marjańska M, Auerbach EJ, et al. In vivo diffusion-weighted MRS using semi-LASER in the human brain at 3 T: methodological aspects and clinical feasibility. NMR Biomed. 2020;e4206. https://doi.org/10.1002/nbm.4206
- editing Deelchand DK, Marjańska M, Henry P-G, Terpstra M. MEGA-PRESS of GABA+: influences of acquisition parameters. NMR Biomed. 2019;e4199. https://doi.org/10.1002/nbm.4199 Ma RE, Murdoch JB, Bogner W, Andronesi O, Dydak U. Atlas-based GABA mapping with 3D MEGA-MRSI: cross-correlation to single-voxel MRS. NMR Biomed. 2020;e4275. https://doi.org/10.1002/nbm.4275 Bell T, Boudes ES, Loo RS, et al. In vivo Glx and Glu measurements from GABA-edited MRS at 3 T. NMR Biomed. 2020;e4245. https://doi.org/10.1002/nbm.4245
- novel acquisition techniques Kulpanovich A, Tal A. What is the optimal schedule for multiparametric MRS? A magnetic resonance fingerprinting perspective. NMR Biomed. 2019;e4196. https://doi.org/10.1002/nbm.4196 Posse S, Sa De La Rocque Guimaraes B, Hutchins-Delgado T, et al. On the acquisition of the water signal during water suppression: high-speed MR spectroscopic imaging with water referencing and concurrent functional MRI. NMR Biomed. 2020;e4261. https://doi.org/10.1002/nbm.4261
- processing techniques Francischello R, Geppi M, Flori A, Vasini EM, Sykora S, Menichetti L. Application of low-rank approximation using truncated singular value decomposition for noise reduction in hyperpolarized 13C NMR spectroscopy. NMR Biomed. 2020;e4285. https://doi.org/10.1002/nbm.4285 Marjańska M, Terpstra M. Influence of fitting approaches in LCModel on MRS quantification focusing on age-specific macromolecules and the spline baseline. NMR Biomed. 2019;e4197. https://doi.org/10.1002/nbm.4197 Landheer K, Swanberg KM, Juchem C. Magnetic resonance Spectrum simulator (MARSS), a novel software package for fast and computationally efficient basis set simulation. NMR Biomed. 2019;e4129. https://doi.org/10.1002/nbm.4129
- reported applications probe the feasibility of novel uses of advanced MRS methods van Houtum Q, Mohamed Hoesein FAA, Verhoeff JJC, et al. Feasibility of 31P spectroscopic imaging at 7 T in lung carcinoma patients. NMR Biomed. 2019;e4204. https://doi.org/10.1002/nbm.4204 Rioux JA, Hewlett M, Davis C, et al. Mapping of fatty acid composition with free-breathing MR spectroscopic imaging and compressed sensing. NMR Biomed. 2019;e4241. https://doi.org/10.1002/nbm.4241 Peeters TH, van Uden MJ, Rijpma A, Scheenen TWJ, Heerschap A. 3D 31P MR spectroscopic imaging of the human brain at 3 T with a 31P receive array: an assessment of 1H decoupling, T1 relaxation times, 1H-31P nuclear Overhauser effects and NAD+. NMR Biomed. 2019;e4169. https://doi.org/10.1002/nbm.4169
Several new publications here on advanced editing techniques - http://www.gabamrs.com/
Voxel placement publication
AutoVOI: real‐time automatic prescription of volume‐of‐interest for single voxel spectroscopy Young Woo Park Dinesh K. Deelchand James M. Joers Brian Hanna Adam Berrington Joseph S. Gillen Kejal Kantarci Brian J.Soher Peter B. Barker HyunWook Park Gülin Öz Christophe Lenglet Magn Reson MEd. 2018;80:1787-1798 https://doi.org/10.1002/mrm.27203
New open-source processing package currently under evaluation -
Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data Georg Oeltzschner Helge J.Zöllner Steve C.N.Hui Mark Mikkelsen Muhammad G.Saleh Sofie Tapper Richard A.E. Edden Journal of Neuroscience Methods Volume 343, 1 September 2020, 108827 https://doi.org/10.1016/j.jneumeth.2020.108827
Presentation at 2020 ENC Meeting - Accelerated MR spectroscopic imaging—a review of current and emerging techniques
Accelerated MR spectroscopic imaging—a review of current and emerging techniques Wolfgang Bogner Ricardo Otazo Anke Henning NMR in Biomedicine.2020;e4314 https://doi.org/10.1002/nbm.4314
Spectroscopy Data Acquisition Tools at CNI
Spectroscopy Sequences
The Stanford CNI effort has become a best practice through a community effort with spectroscopy expertise from CNI staff, MRI scientists, and an expanding user community. Current spectroscopy sequences include methods both for edited GABA (gamma-Aminobutyric acid) specific data acquisition (MEGA-PRESS [1] and IM-SPECIAL [3]), and for multi-metabolite data acquisition (Optimized-PRESS [4], [5], [6]) and semi-LASER [9], [10], [11], [12], [13] Most sequences are linked to automatic processing pipelines in CNI's data management system, Flywheel, as referenced in the table below.
| Spectroscopy Sequence | Measured Metabolites | Analysis Methods |
|---|---|---|
| MEGA-PRESS [1] | GABA+, Glx(Glutamate, Glutmine) | Gannet [2] |
| IM-SPECIAL [3] | GABA, Glu(Glutamate), Glx(Glutamate, Glutamine) | Sequence specific Matlab code |
| Optimized-PRESS [4], [5], [6] | All metabolites | LCModel fitting [7] |
| semi-LASER [9], [10], [11], [12], [13] | All metabolites | LCModel fitting [7] |
References
[1] Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R (1998) Simultaneous in vivo spectral editing and water suppression, NMR Biomed. 11:266–272 https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291099-1492%28199810%2911%3A6%3C266%3A%3AAID-NBM530%3E3.0.CO%3B2-J
[2] Richard A.E. Edden, Nicolaas A.J. Puts, Ashley D. Harris, Peter B. Barker, and C. John Evans (2014) Gannet: A Batch-Processing Tool for the Quantitative Analysis of Gamma-Aminobutyric Acid–Edited MR Spectroscopy Spectra, Journal of Magnetic Resonance Imaging 40:1445–1452 https://doi.org/10.1002/jmri.24478
[3] Gu M, Hurd R, Noeske R, Baltusis L, Hancock R, Sacchet MD, Gotlib IH, Chin FT, Spielman DM (2018) GABA editing with macromolecule suppression using an improved MEGA-SPECIAL sequence, Magnetic Resonance in Medicine 79:41-47 https://doi.org/10.1002/mrm.26691
[4] Webb PG, Sailasuta N, Kohler SJ, Raidy T, Moats RA, Hurd R (1994) Automated single voxel proton MRS: technical development and multisite verification, Magnetic Resonance in Medicine 31(4):365-373 https://doi.org/10.1002/mrm.1910310404
[5] Bodenhausen G, Freeman R, Turner DL (1977) Suppression of artifacts in two dimensional J spectroscopy, Journal of Magnetic Resonance Imaging 27:511-514 https://doi.org/10.1016/0022-2364(77)90016-6
[6] Tran TK, Vigneron DB, Sailasuta N, Tropp J, Le Roux P, Kurhanewicz J, Nelson S, Hurd R (2000) Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer, Magnetic Resonance in Medicine 43(1):23-33. https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291522-2594%28200001%2943%3A1%3C23%3A%3AAID-MRM4%3E3.0.CO%3B2-E
[7] Provencher SW (2001) Automatic quantitation of localized in vivo1H spectra with LCModel, NMR in Biomedicine 14(4):260-264 https://doi.org/10.1002/nbm.698
[8] Young Woo Park, Dinesh K. Deelchand, James M. Joers, Brian Hanna, Adam Berrington, Joseph S. Gillen, Kejal Kantarci, Brian J. Soher, Peter B. Barker, HyunWook Park, Gulin Oz, Christophe Lenglet (2018) AutoVOI: real-time automatic prescription of volume-of-interest for single voxel spectroscopy, Magn. Reson. Med. 80:1787–1798 https://doi.org/10.1002/mrm.27203
[9] Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A. (2018) Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses, Magn. Reson. Med. 59(1):1-6 https://doi.org/10.1002/mrm.21302
[10] Oz G, Tkac I. (2011) Short-echo, single-shot, full-intensity proton magnetic resonance spectroscopy for neurochemical profiling at 4 T: validation in the cerebellum and brainstem, Magn. Reson. Med. 65(4):901-910 https://doi.org/10.1002/mrm.22708
[11] Martin Wilson, Ovidiu Andronesi, Peter B. Barker, Robert Bartha, Alberto Bizzi, Patrick J. Bolan, Kevin M. Brindle, In-Young Choi, Cristina Cudalbu, Ulrike Dydak, Uzay E. Emir, Ramon G. Gonzalez, Stephan Gruber, Rolf Gruetter, Rakesh K. Gupta, Arend Heerschap, Anke Henning,Hoby P. Hetherington, Petra S. Huppi, Ralph E. Hurd, Kejal Kantarci, Risto A Kauppinen, Dennis W. J. Klomp, Roland Kreis, Marijn J. Kruiskamp, Martin O. Leach, Alexander P. Lin, Peter R. Luijten, Malgorzata Marjanska, Andrew A. Maudsley, Dieter J. Meyerhoff, Carolyn E. Mountford, Paul G. Mullins, James B. Murdoch, Sarah J. Nelson, Ralph Noeske, Gulin Oz, Julie W. Pan, Andrew C. Peet, Harish Poptani, Stefan Posse, Eva-Maria Ratai, Nouha Salibi, Tom W. J. Scheenen, Ian C. P. Smith, Brian J. Soher, Ivan Tkac, Daniel B. Vigneron, Franklyn A. Howe (2019) Methodological consensus on clinical proton MRS of the brain:Review and recommendations, Magn. Reson. Med. 82:527–550 https://doi.org/10.1002/mrm.27742
[12] Dinesh K. Deelchand, Adam Berrington, Ralph Noeske, James M. Joers, Arvin Arani, Joseph Gillen, Michael Schar, Jon-Fredrik Nielsen, Scott Peltier, Navid Seraji-Bozorgzad, Karl Landheer, Christoph Juchem, Brian Soher, Douglas C. Noll, Kejal Kantarci, Eva M. Ratai, Thomas H. Marecii, Peter B. Barker, Gulin Oz (2019) Across-vendor standardization of semi-LASER for single-voxel MRS at 3T, NMR in Biomedicine. e4218 https://doi.org/10.1002/nbm.4218
[13] Gulin Oz, Dinesh K. Deelchand, Jannie P. Wijnen, Vladimir Mlynarik, Lijing Xin, Ralf Mekle, Ralph Noeske, Tom W.J. Scheenen, Ivan Tkac, the Experts' Working Group on Advanced Single Voxel 1H MRS (2020) Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: Experts' consensus recommendations, NMR in Biomedicine. e4236 https://doi.org/10.1002/nbm.4236
Current CNI spectroscopy protocols
There are two CNI spectroscopy protocols
(1) The protocol spectro-protocol-1 located in the CNI "Other" tab on the scanner has the sLaser and optimized-PRESS sequences for data collection for all metabolites. For new studies users should use the sLaser sequence. For longitudinal studies the optimized-PRESS sequence can be continued to be used.
(2) The protocol spectro-protocol-1 - editing located in the CNI "Other" tab on the scanner has the MEGA-PRESS sequence for data collection for GABA.
Protocol screen shot: Spectro-protocol-1

| Spectroscopy Sequence | Description | Parameters |
|---|---|---|
| Localizer | ||
| T1 | ||
| T2 | ||
| Voxel Placement | ||
| s-Laser | This sequence is the GE "Works in Progress" and is consistent with the ISMRM Experts' consensus recommendations for this sequence. "High field (3T) and ultra-high field (7T) are ideal field strength for MR spectroscopy due to the higher spectral resolution and higher signal that can be achieved. But with these advantages comes a higher B1-inhomogeneity and larger Chemical Shift Displacement Error (CSDE). Semi-LASER is a double spin-echo MRS technique like the established PRESS (GE product name Probe-P) technique that uses a slice selective non-adiabatic excitation and two pairs of adiabatic slice selective refocusing pulses for volume selection. The adiabatic behavior of the RF pulses addresses the B1-inhomogeneity problem while the increased bandwidth of these pulses reduces the CSDE." | |
| Optimized-PRESS | This sequence is a GE "Works in Progress" and is consistent with the ISMRM Experts' consensus recommendations for this sequence. This is currently considered a legacy sequence. |
Screen shots of parameters for s-laser sequence
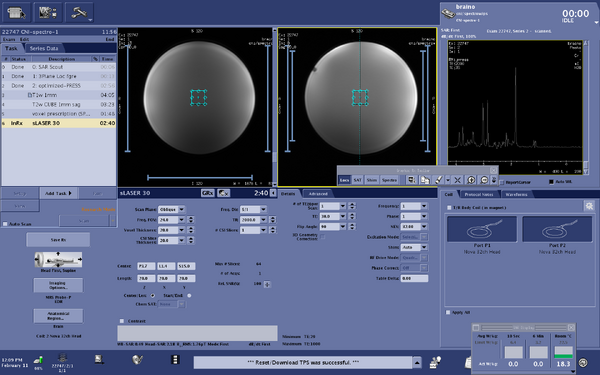 |
 |
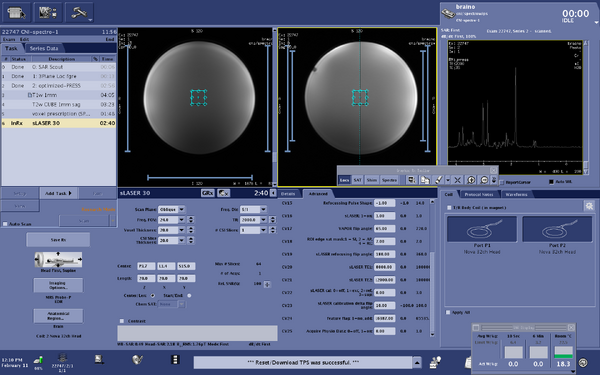 |
Screen shots of parameters for optimized-press sequence
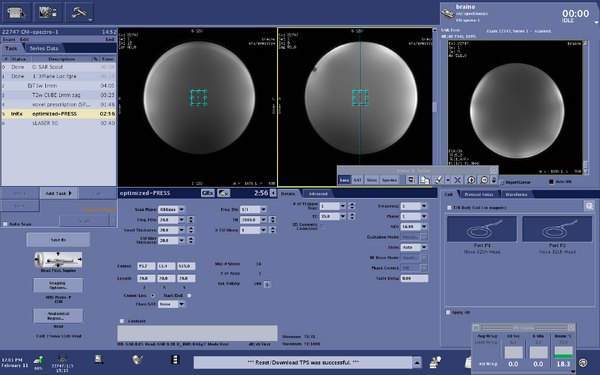 |
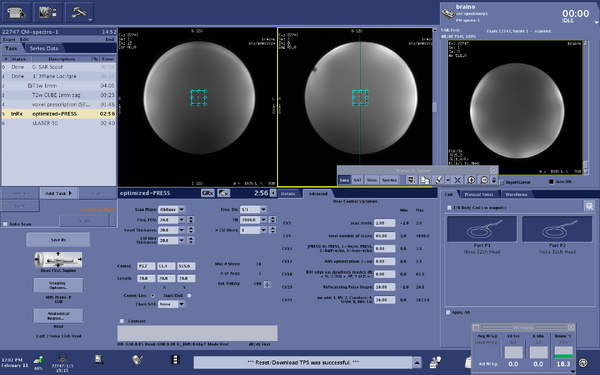 |
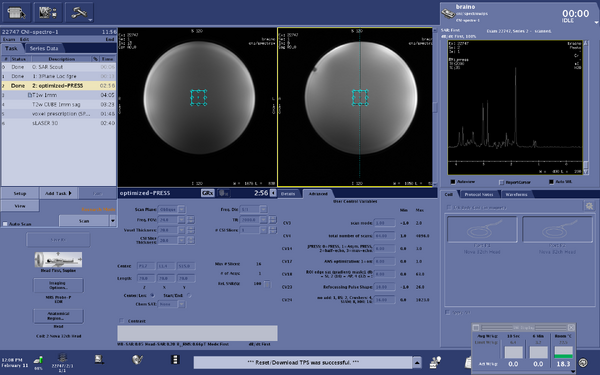 |
Protocol screen shot:Spectro-protocol-editing-1
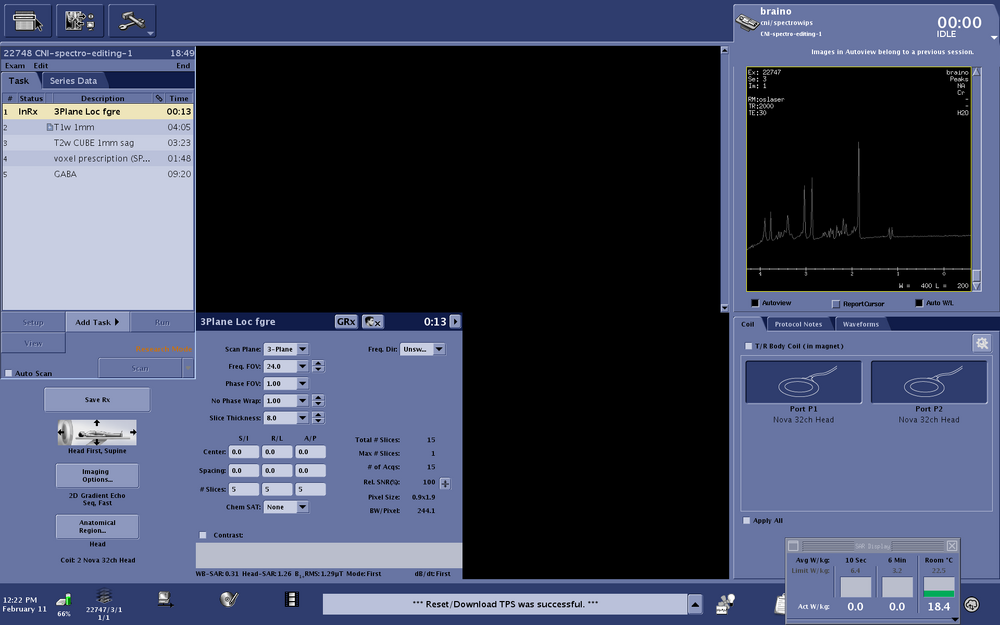
| Spectroscopy Sequence | Description | Parameters |
|---|---|---|
| Localizer | ||
| T1 | ||
| T2 | ||
| Voxel Placement | ||
| MEGA-PRESS | The current spectroscopy sequence for edited GABA (gamma-Aminobutyric acid) data acquisition is MEGA-PRESS. This sequence is the GE "Works in Progress" and is consistent with the ISMRM Experts' consensus recommendations for this sequence. The only modification of the CNI version from the GE Works in progress is the value of TR (CNI TR = 2000, changed from TR = 1800)
MEGA-PRESS description for publications - Example MEGA-PRESS description for publication from Association between anterior cingulate neurochemical concentration and individual differences in hypnotizability DeSouza DD, Stimpson K, Baltusis L, Sacchet MD, Gu M, Hurd R, Wu H, Yeomans DC, Williams N, Spiegel D. Association between anterior cingulate neurochemical concentration and individual differences in hypnotizability. Cerebral Cortex, Volume 30, Issue 6, June 2020, Pages 3644–3654, https://doi.org/10.1093/cercor/bhz332, Additional example sequence description from GABA editing with macromolecule suppression using an improved MEGA-SPECIAL sequence Gu M, Hurd R, Noeske R, Baltusis L, Hancock R, Sacchet MD, Gotlib IH, Chin FT, Spielman DM (2018) GABA editing with macromolecule suppression using an improved MEGA-SPECIAL sequence, Magnetic Resonance in Medicine 79:41-47 https://doi.org/10.1002/mrm.26691 |
MRI system description
Acquisition parameters
|
 |
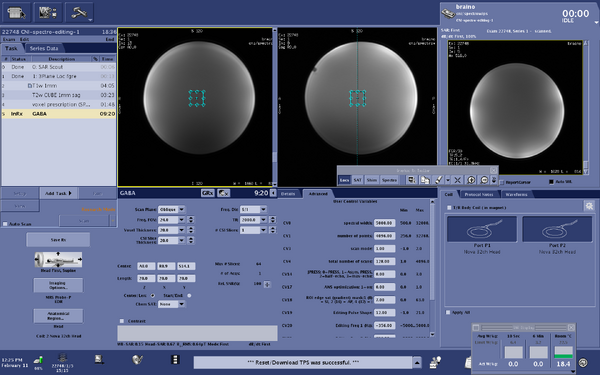 |
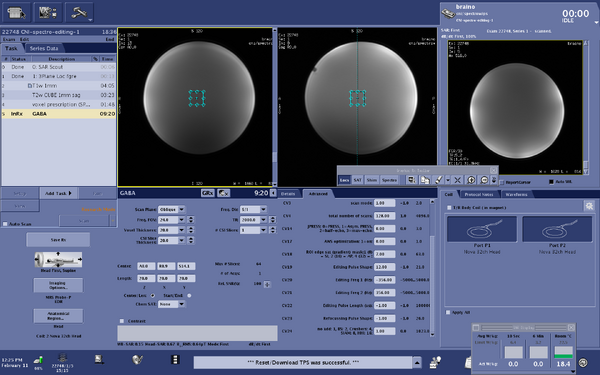 |
 |
 |
Prescribing a rotated voxel
At the moment GE’s DV26 and higher software platforms release doesn’t support voxel rotation for spectroscopy sequences. A workaround however is available:
(1) After the 3-plane reconstruction of the T1w images, setup a 3D imaging scan. We saved a template of this 3D scan in the protocol “CNI Example Spectroscopy” and named it as “Voxel prescription”. Add this sequence to your protocol and put it before the MRS scan.
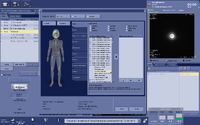
(2) Setup the voxel prescription scan. The Scan Plane is set to “Oblique”, FOV and Locs per Slab are set as close to the voxel size of the spectro scan as possible. Prescribe the 3D box, place it to the desired location and rotate to the right angle. Because the box is usually bigger than the voxel size you want to prescribe, the coverage you see in this step is not accurate. It’s crucial to get the correct orientation of the box. Save Rx.
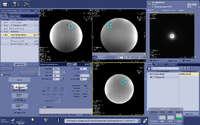
(3) Setup the MRS scan. Copy Rx from the voxel prescription scan. By default the Mode Filter in the Copy Rx is set to “MRS”, you need to change it to “All” or “3D” in order to see the voxel prescription scan in the Copy Rx list. Select it and accept.
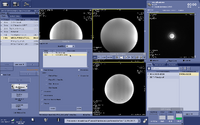
(4) Adjust voxel location and size. You can move the box around and change its size on the graphic interface, or set its coordinates and size by setting the Center and Length in X, Y, Z. If you want to adjust the orientation of the voxel, you need to go back to the 3D voxel prescription scan, and repeat step 2-4 again. After you finish setting up the MRS scan, Save Rx and proceed with scanning.

Extracting voxel prescription information retrospectively
The prescription information of the voxel of an MRS scan is saved in the header of the raw data (p-file). Specifically, the x, y, z dimensions of the voxel are stored in header fields rdb_hdr_image.user8, rdb_hdr_image.user9, rdb_hdr_image.user10; the x, y, z locations of the center of the voxel are stored in header fields rdb_hdr_image.user11, rdb_hdr_image.user12, rdb_hdr_image.user13, in RAS coordinates (e.g. if the voxel is at L26.0 then rdb_hdr_image.user11 will record -26.0). The unit is mm.
To look for this information on Flywheel, open the information window of the p-file, and search for op_user_8, op_user_9, op_user_10, etc.
Spectroscopy Data Processing Tools at CNI
Data Management
CNI currently uses Flywheel as its data base management system. A critical feature of this data base management is the ability to share computational methods within the system. The CNI now provides a combination of data repository and integrated open source processing tools such as Gannet [2] and LCModel [7]. This combination of tools supports scientific transparency for both data and computational sharing. Spectroscopy analysis methods such as LCModel can be containerized as a gear in Flywheel for automated processing and data visualization.
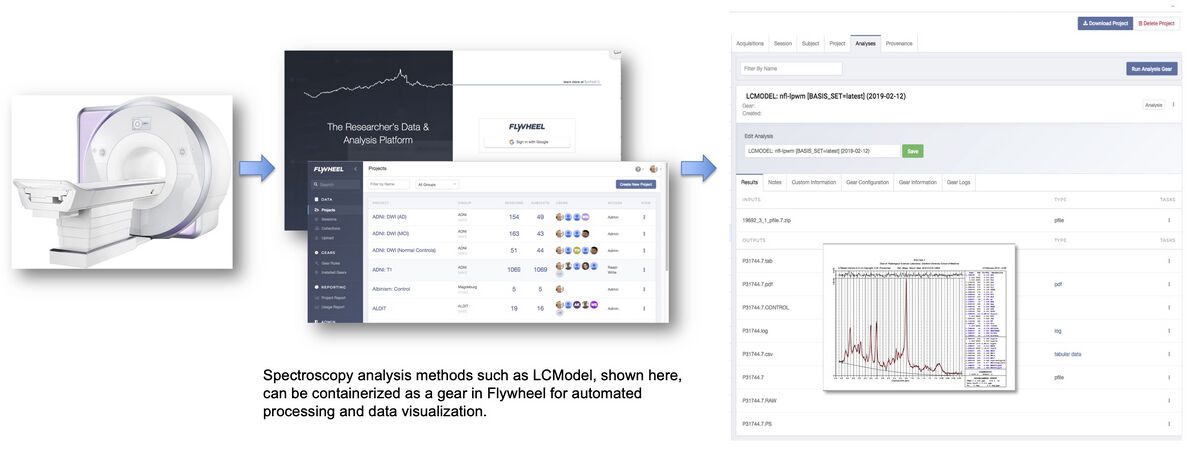
Current CNI spectroscopy Flywheel gears
Each of the supported spectroscopy sequences is linked to a Flywheel gear for automatic data processing
| Spectroscopy Sequence | Measured Metabolites | Analysis Methods |
|---|---|---|
| MEGA-PRESS | GABA+, Glx(Glutamate, Glutmine) | Gannet Flywheel processing gear: Gannet 3.1 Description and Figures here -
|
| Optimized-PRESS | All metabolites | Sequence specific Matlab code, LCModel fitting - Flywheel processing gear information here: LCMODEL_r_dlpfc_nfl_[BASIS_SET=02_01_19,FIXED_NAA=0]_(2021-05-13) |
| semi-LASER | All metabolites | Sequence specific Matlab code, LCModel fitting - Flywheel processing gear information here: LCMODEL_r_dlpfc_nfl_[BASIS_SET=02_01_19,FIXED_NAA=0]_(2021-05-13) |
Location of data analysis results in Flywheel
Note: For users using the CNI Optimized PRESS "nfl" sequence with LCModel data analysis - Users who are in groups that are not themselves Flywheel "Lab" customers will need to download their results from the session tab now (not the analyses tab as was done before and shown in the figure below)
If you are using Google Chrome as the browser to download the processed data, then you will need to add the extension .zip to the downloaded file name. This operation does not not need to be done if using Safari or Firefox browsers.
MEGA-PRESS results will continue to be in the analyses tab.
