Historical Information: Difference between revisions
imported>Laimab No edit summary |
imported>Laimab No edit summary |
||
| Line 205: | Line 205: | ||
|[[File:SPECIAL7.png|200px|thumb|left|SPECIAL7]] | |[[File:SPECIAL7.png|200px|thumb|left|SPECIAL7]] | ||
|} | |} | ||
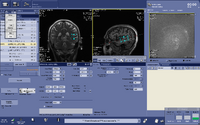
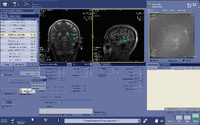
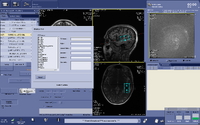
Automated voxel placement tools: | |||
Typically MRS data is collected within a single voxel that needs to be manually prescribed. Manual prescription can lead to systematic bias and low data quality that may be caused by inexperienced users or inter-individual placement variability. To avoid these issues, and improve data collection quality, Matthew Sacchet has developed an automated voxel placement tool. It uses non-linear warping between native subject space and template space to identify precise voxel locations in scanner space. The tool is used in real-time during data acquisition and has been streamlined for efficient usage and low time cost. | |||
Revision as of 01:08, 1 April 2021
Older methods and literature references will go here...
Extra section to be removed
Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann NY Acad Sci 1987;508:333–48 https://doi.org/10.1111/j.1749-6632.1987.tb32915.x
 |
 |
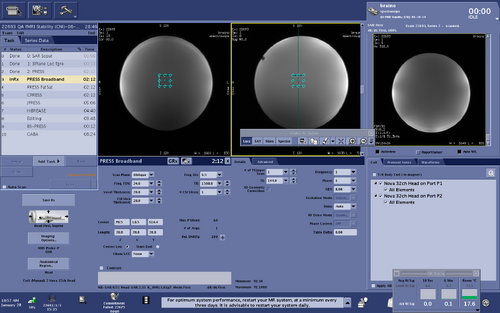
PRESS Broadband
The product refocusing pulses within PRESS can be replaced by broadband refocusing pulses described in M. Janich et al., Slice-selective broadband refocusing pulses for the robust generation of crushed spin-echoes, J Magn Reson, 223: 129 – 137 (2012) https://doi.org/10.1016/j.jmr.2012.08.003 to reduce the CSDE.
 |
 |
CV 0 5000
CV0: spectral width The total spectral frequency width in Hz.
CV 1 4096
CV1: number of points The number of complex data points acquired per excitation.
CV 4 128 (1.5T) 64 (3T)
CV4: total number of scans The total number of excitations acquired per echo (cycle) when signal averaging. The value of CV4 must be an integer multiple of NEX.
CV 14 0
CV14: JPRESS Mode
0 (PRESS) standard PRESS acquisition, TE1 is set by sequence to minimum
1 (asym. PRESS) TE1 is set by user
2 (half-echo) standard JPRESS acquisition mode, echo acquired from echo top
3 (max-echo) maximum-echo mode described in [6]. For shortest echo time echo is acquired from echo top, for longer echo times data acquisition starts before the echo top.
CV 18 7
CV18: ROI edge sat/gradient mask There are three pairs of VSS RF pulses available at the edge of the Spectroscopy VOI. The VSS can be selected or deselected.
0 – no VSS RF pulses;
1 – S/I the superior and inferior pulses only;
2 – A/P the anterior and posterior pulses only;
3 – S/I and A/P, two pulse pairs;
4 – R/L the right and the left pulses only;
5 – R/L and S/I, two pulse pairs;
6 - R/L and A/P, two pulse pairs;
7 – R/L, A/P and S/I, three pulse pairs, this is the default value
This bitmask determines the polarity of the slice gradients of the three PRESS excitation/refocusing pulses and therefore the direction of the fat-water chemical shift. The corresponding bit set to 1 is negative polarity, 0 is positive.
0 – S/I, A/P and R/L all positive, this is the default value
8 – S/I negative, A/P and R/L positive
16 – A/P negative, S/I and R/L positive
24 – S/I and A/P negative, R/L positive
32 – R/L negative, S/I and A/P positive
40 – R/L and S/I negative, A/P positive
48 - R/L and A/P negative, S/I positive
56 – R/L, A/P and S/I all negative
CV 23 20
CV23: Refocusing Pulse Shape: PRESS refocusing pulse shape
20 – S-BREBOP-7500.rho, pw = 7.5ms
26 – flip_1803.rho, non-linear phase refocusing pulse used for CPRESS
-1 – default pulse set by PSD
CV 24 1
CV24: Feature flag
1 (no add) Store each single data acquisition in pfile.
2 (Bloch-Siegert) Bloch-Siegert TG calibration
PRESS FatSat
The dual BASING technique can be used for additional metabolite suppression, e.g. fat.
J. Star-Lack et al., Improved Water and Lipid Suppression for 3D PRESS CSI Using RF Band Selective Inversion with Gradient Dephasing (BASING), Magn Reson Med, 38, 311 – 321, (1997) https://doi.org/10.1002/mrm.1910380222
CPRESS
CPRESS is a modified PRESS spectroscopic localization pulse sequence that replaces each of the two refocusing RF pulses with a pair of non-linear phase refocusing pulses. The non-linear phase refocusing pulses has been designed to operate at a lower maximum B1 requirement (0.15 G) while keeping the pulse width short (4.3 ms) and maintaining adequate bandwidth for spectroscopy application at 3T (1.2 kHz). At a TE of 42 ms, the short inter pulse delay (10.5 ms) between the refocusing pulses has a potential to provide improved sensitivity for J-coupled metabolites such as myoinositol (mI) and glutamate / glutamine (Glx) over conventional short TE (35 ms) PRESS sequence.
I. Hancu, Which pulse sequence is optimal for myo-inositol detection at 3T? NMR Biomed, 22: 426 – 435 (2009) https://doi.org/10.1002/nbm.1353
I. Hancu et al, Improved Myo-inositol Detection Through Carr–Purcell PRESS: A Tool for More Sensitive Mild Cognitive Impairment Diagnosis, Magn Reson Med, 65: 1515 – 1521 (2011) https://doi.org/10.1002/mrm.22749
JPRESS
The two-dimensional J-resolved spectroscopy sequence consists of a series of spin-echo experiments with different echo times defined by the average echo time (TE) and the delta of echo times (User CV 16) employing PRESS localization. Adding the spectra gives a TEA-PRESS spectrum.
L. Ryner et al., Localized 2D J-resolved H-1 MR spectroscopy – strong coupling effects in vitro and in vivo, Magn Reson Imaging, 13: 853 – 869 (1995) https://doi.org/10.1016/0730-725x(95)00031-b
R. Schulte, Improved two-dimensional J-resolved spectroscopy, NMR Biomed, 19: 264 – 270 (2006) https://doi.org/10.1002/nbm.1027
mBREASE
mBREASE is a TEA-PRESS sequence with enabled STIR fat suppression, storage of each aquired FID to correct for frequency shifts and acquisition of reference data with different echo times to correct for water T2. It includes quantification of the choline signal using a voigt-lineshape model function in time domain and linear baseline in frequency domain. T2 corrected water signal is used as internal reference.
Editing
Spectral j-difference editing describes an advanced spectroscopy acquisition technique which is generally necessary to detect j-coupled markers and separate from co-resonant metabolite peaks. The implemented editing technique is based on the BASING technique described.
J. Star-Lack et al., Improved Water and Lipid Suppression for 3D PRESS CSI Using RF Band Selective Inversion with Gradient Dephasing (BASING), Magn Reson Med, 38, 311 – 321, (1997) https://doi.org/10.1002/mrm.1910380222
J. Star-Lack et al., In Vivo Lactate Editing with Simultaneous Detection of Choline, Creatine, NAA, and Lipid Singlets at 1.5 T Using PRESS Excitation with Applications to the Study of Brain and Head and Neck Tumors, J Magn Reson, 133: 243 – 254 (1998) https://doi.org/10.1006/jmre.1998.1458
BS-PRESS
BS-PRESS is a voxel based TG calibration method described in [8]. The phase-based B1+ mapping technique using the Bloch-Siegert shift method encodes the B1 information into a signal phase resulting from off-resonant RF pulses within the sequence [9-11].
8. R. Noeske et al., Voxel Based Transmit Gain Calibration using Bloch-Siegert Shift Method for MR Spectroscopy, Proc 20th Annual Meeting ISMRM, Melbourne: 1733 (2012)
9. Sacolick et al., B1 Mapping by Bloch-Siegert Shift, Magn Reson Med, 63: 1315 - 1322 (2010)
10. Sacolick et al., Fast Radiofrequency Flip Angle Calibration by Bloch–Siegert Shift, Magn Reson Med, 66: 1333 - 1338 (2011)
11. Sacolick et al., Fast Spin Echo Bloch-Siegert B1 Mapping, Proc 19th Annual Meeting ISMRM, Montreal: 2927 (2011)
GABA
Older protocols (and sequences):
The CNI GABA spectroscopy MEGA PRESS protocol (CNI 32ch Participant GABA Spectroscopy) (located in CNI/Head), which can be run independently or added to an fMRI protocol.
There is now also the CNI-PRESS-SPECIAL (located in CNI/Head) protocol for GABA data collection, which can be run independently or added to an fMRI protocol. This is the latest protocol and the one to use.
Key Parameters for sequences:
This section (in the process of being updated) includes:
- parameters that require adjustment or checking prior to collecting spectroscopy data.
- additional setup sequences prior to running the spectroscopy sequence (example - shims) with setup details.
 |
 |
 |
 |
 |
 |
 |
 |
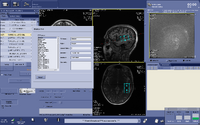 |
Automated voxel placement tools:
Typically MRS data is collected within a single voxel that needs to be manually prescribed. Manual prescription can lead to systematic bias and low data quality that may be caused by inexperienced users or inter-individual placement variability. To avoid these issues, and improve data collection quality, Matthew Sacchet has developed an automated voxel placement tool. It uses non-linear warping between native subject space and template space to identify precise voxel locations in scanner space. The tool is used in real-time during data acquisition and has been streamlined for efficient usage and low time cost.