Spectro
MRS in Neurosciences: In-vivo spectroscopy methods and applications at CNI
Overview
Discoveries about the brain have implications for fields ranging from Business, Law, Psychology, and Education. The interest of measuring metabolic changes via MRS techniques and combining that information with functional MRI measurements continues to grow. CNI continues to support the research of its user community by providing state-of-the art data acquisition and state-of-the-art data management and analysis capabilities for in-vivo spectroscopy. Through collaborative efforts the special interest spectroscopy group at CNI has enabled education, participated in experimental design, and guided analyses and interpretation of results.
An excellent spectroscopy resource website is https://mrshub.org/
The MRSHub is a curated collection of resources for the analysis of magnetic resonance spectroscopy data. It is maintained by the Committee for MRS Code and Data Sharing of the MR Spectroscopy Study Group of the International Society for Magnetic Resonance in Medicine (ISMRM).
Data Acquisition and Processing Tools
Spectroscopy Sequences
The Stanford CNI effort has become a best practice through a community effort with spectroscopy expertise from CNI staff, MRI scientists, and an expanding user community. Current spectroscopy sequences include methods both for edited GABA (gamma-Aminobutyric acid) specific data acquisition (MEGA-PRESS [1] and IM-SPECIAL [3]), and for multi-metabolite data acquisition (Optimized-PRESS [4], [5], [6]). Additional sequences such as semi-LASER [9], [10], [11], [12], [13] are being evaluated and added as newer data acquisition methods.
| Spectroscopy Sequence | Measured Metabolites | Analysis Methods |
|---|---|---|
| MEGA-PRESS [1] | GABA+, Glx(Glutamate, Glutmine) | Gannet [2] |
| IM-SPECIAL [3] | GABA, Glu(Glutamate), Glx(Glutamate, Glutamine) | Sequence specific Matlab code |
| Optimized-PRESS [4], [5], [6] | All metabolites | Sequence specific Matlab code, LCModel fitting [7] |
| semi-LASER [9], [10], [11], [12], [13] | All metabolites | Sequence specific Matlab code, LCModel fitting [7] |
References
[1] Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R (1998) Simultaneous in vivo spectral editing and water suppression, NMR Biomed. 11:266–272 https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291099-1492%28199810%2911%3A6%3C266%3A%3AAID-NBM530%3E3.0.CO%3B2-J
[2] Richard A.E. Edden, Nicolaas A.J. Puts, Ashley D. Harris, Peter B. Barker, and C. John Evans (2014) Gannet: A Batch-Processing Tool for the Quantitative Analysis of Gamma-Aminobutyric Acid–Edited MR Spectroscopy Spectra, Journal of Magnetic Resonance Imaging 40:1445–1452 https://doi.org/10.1002/jmri.24478
[3] Gu M, Hurd R, Noeske R, Baltusis L, Hancock R, Sacchet MD, Gotlib IH, Chin FT, Spielman DM (2018) GABA editing with macromolecule suppression using an improved MEGA-SPECIAL sequence, Magnetic Resonance in Medicine 79:41-47 https://doi.org/10.1002/mrm.26691
[4] Webb PG, Sailasuta N, Kohler SJ, Raidy T, Moats RA, Hurd R (1994) Automated single voxel proton MRS: technical development and multisite verification, Magnetic Resonance in Medicine 31(4):365-373 https://doi.org/10.1002/mrm.1910310404
[5] Bodenhausen G, Freeman R, Turner DL (1977) Suppression of artifacts in two dimensional J spectroscopy, Journal of Magnetic Resonance Imaging 27:511-514 https://doi.org/10.1016/0022-2364(77)90016-6
[6] Tran TK, Vigneron DB, Sailasuta N, Tropp J, Le Roux P, Kurhanewicz J, Nelson S, Hurd R (2000) Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer, Magnetic Resonance in Medicine 43(1):23-33. https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291522-2594%28200001%2943%3A1%3C23%3A%3AAID-MRM4%3E3.0.CO%3B2-E
[7] Provencher SW (2001) Automatic quantitation of localized in vivo1H spectra with LCModel, NMR in Biomedicine 14(4):260-264 https://doi.org/10.1002/nbm.698
[8] Young Woo Park, Dinesh K. Deelchand, James M. Joers, Brian Hanna, Adam Berrington, Joseph S. Gillen, Kejal Kantarci, Brian J. Soher, Peter B. Barker, HyunWook Park, Gulin Oz, Christophe Lenglet (2018) AutoVOI: real-time automatic prescription of volume-of-interest for single voxel spectroscopy, Magn. Reson. Med. 80:1787–1798 https://doi.org/10.1002/mrm.27203
[9] Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A. (2018) Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses, Magn. Reson. Med. 59(1):1-6 https://doi.org/10.1002/mrm.21302
[10] Oz G, Tkac I. (2011) Short-echo, single-shot, full-intensity proton magnetic resonance spectroscopy for neurochemical profiling at 4 T: validation in the cerebellum and brainstem, Magn. Reson. Med. 65(4):901-910 https://doi.org/10.1002/mrm.22708
[11] Martin Wilson, Ovidiu Andronesi, Peter B. Barker, Robert Bartha, Alberto Bizzi, Patrick J. Bolan, Kevin M. Brindle, In-Young Choi, Cristina Cudalbu, Ulrike Dydak, Uzay E. Emir, Ramon G. Gonzalez, Stephan Gruber, Rolf Gruetter, Rakesh K. Gupta, Arend Heerschap, Anke Henning,Hoby P. Hetherington, Petra S. Huppi, Ralph E. Hurd, Kejal Kantarci, Risto A Kauppinen, Dennis W. J. Klomp, Roland Kreis, Marijn J. Kruiskamp, Martin O. Leach, Alexander P. Lin, Peter R. Luijten, Malgorzata Marjanska, Andrew A. Maudsley, Dieter J. Meyerhoff, Carolyn E. Mountford, Paul G. Mullins, James B. Murdoch, Sarah J. Nelson, Ralph Noeske, Gulin Oz, Julie W. Pan, Andrew C. Peet, Harish Poptani, Stefan Posse, Eva-Maria Ratai, Nouha Salibi, Tom W. J. Scheenen, Ian C. P. Smith, Brian J. Soher, Ivan Tkac, Daniel B. Vigneron, Franklyn A. Howe (2019) Methodological consensus on clinical proton MRS of the brain:Review and recommendations, Magn. Reson. Med. 82:527–550 https://doi.org/10.1002/mrm.27742
[12] Dinesh K. Deelchand, Adam Berrington, Ralph Noeske, James M. Joers, Arvin Arani, Joseph Gillen, Michael Schar, Jon-Fredrik Nielsen, Scott Peltier, Navid Seraji-Bozorgzad, Karl Landheer, Christoph Juchem, Brian Soher, Douglas C. Noll, Kejal Kantarci, Eva M. Ratai, Thomas H. Marecii, Peter B. Barker, Gulin Oz (2019) Across-vendor standardization of semi-LASER for single-voxel MRS at 3T, NMR in Biomedicine. e4218 https://doi.org/10.1002/nbm.4218
[13] Gulin Oz, Dinesh K. Deelchand, Jannie P. Wijnen, Vladimir Mlynarik, Lijing Xin, Ralf Mekle, Ralph Noeske, Tom W.J. Scheenen, Ivan Tkac, the Experts' Working Group on Advanced Single Voxel 1H MRS (2020) Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: Experts' consensus recommendations, NMR in Biomedicine. e4236 https://doi.org/10.1002/nbm.4236
[14] L. Ryner, J Sorenson, M.A. Thomas (1995) Localized 2D J-resolved H-1 MR spectroscopy: Strong coupling effects in vitro and in vivo, Magn. Reson. Imaging 13:853–869 https://doi.org/10.1016/0730-725X(95)00031-B
[15] R. Schulte, T. Lange, J. Beck, D. Meier, P. Boesiger (2006) Improved two-dimensional J-resolved spectroscopy, NMR Biomed. 19:264–270 https://doi.org/10.1002/nbm.1027
[16] Krish Krishnamurthy (2013) CRAFT (complete reduction to amplitude frequency table) – robust and time efficient Bayesian approach for quantitative mixture analysis by NMR, Magn. Reson. Chem. 51: 821–829 https://doi.org/10.1002/mrc.4022
Automated magnetic resonance spectroscopy voxel placement tools
Typically MRS data is collected within a single voxel that needs to be manually prescribed. To improve data collection quality, there has been anincreased interest and development of real-time single voxel automated prescription placement methods8. Researchers can now acquire MRS data in a routine way using a variety of automated voxel placement procedures. The procedures are used in real-time during data acquisition and have been streamlined for efficient usage and low time cost.
- One procedure uses non-linear warping between native subject space and template space (i.e., Montreal Neurological Institute [MNI] space) to identify precise voxel locations in scanner space that are based on MNI neuroanatomy.
- A second procedure is for the real-time in-scan session identification of precise coordinates from prior scanning sessions for MRS voxel placement based on person-specific coordinates. For example, if an initial scan session includes a functional task, information from that data can be used for subsequent voxel placement.
- A third procedure that is similar to second procedure above is using the central coordinate of an MRS voxel collected previously for placement in the same individual during a second scan session.
Data Management
CNI currently uses Flywheel as its data base management system. A critical feature of this data base management is the ability to share computational methods within the system. The CNI now provides a combination of data repository and integrated open source processing tools such as Gannet [2] and LCModel [7]. This combination of tools supports scientific transparency for both data and computational sharing. Spectroscopy analysis methods such as LCModel can be containerized as a gear in Flywheel for automated processing and data visualization.
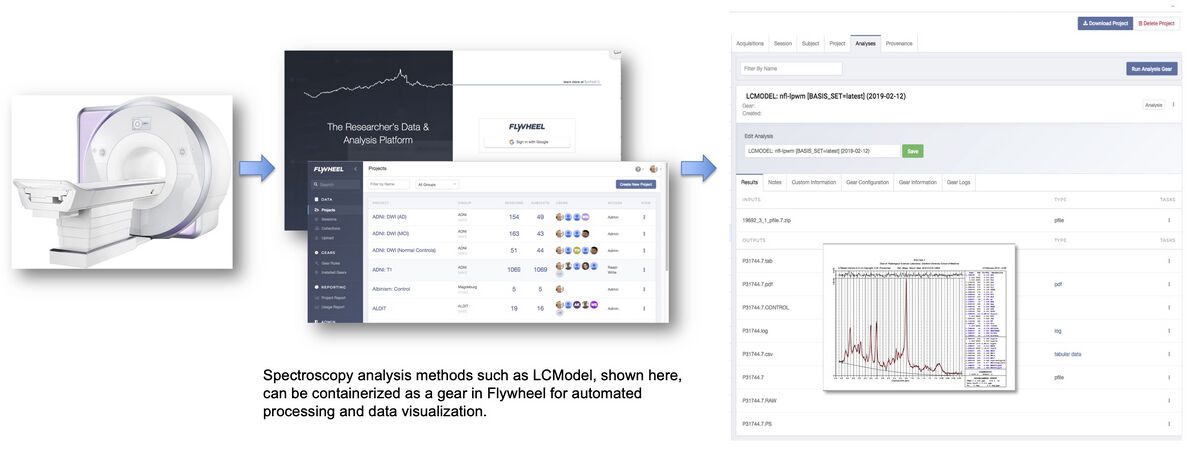
Representative Research Studies
Examples of studies at CNI using in-vivo spectroscopy techniques include characterization of biomarkers following transcranial magnetic stimulation, and metabolite characterization for conditions such as substance addiction, pain, depression, and various forms of dementia. These studies illustrate integrated data acquisition and data processing tools accessible to the CNI user community which simplify the use and sharing of spectroscopy results in neuroimaging applications.
Association between anterior cingulate neurochemical concentration and individual differences in hypnotizability DeSouza DD, Stimpson K, Baltusis L, Sacchet MD, Gu M, Hurd R, Wu H, Yeomans DC, Williams N, Spiegel D. Association between anterior cingulate neurochemical concentration and individual differences in hypnotizability. Cerebral Cortex, Volume 30, Issue 6, June 2020, Pages 3644–3654, https://doi.org/10.1093/cercor/bhz332
New Methods and Techniques
The CNI continues to support the research of its user community by developing and incorporating for general use new data acquisition and data analysis capabilities.
GABA editing with macromolecule suppression using an improved MEGA-SPECIAL sequence Gu M, Hurd R, Noeske R, Baltusis L, Hancock R, Sacchet MD, Gotlib IH, Chin FT, Spielman DM (2018) GABA editing with macromolecule suppression using an improved MEGA-SPECIAL sequence, Magnetic Resonance in Medicine 79:41-47 https://doi.org/10.1002/mrm.26691
Spectroscopy Literature
Recent Literature
Papers presented at the ISMRM MR Spectroscopy Study Group Virtual Meeting: NMR in Biomedicine Special Issue Experts' Recommendations: Where Consensus Was Reached and Where Dissent Prevailed, Part 1 (November 12, 2020)
Advanced methodology for in vivo magnetic resonance spectroscopy In‐Young Choi Roland Kreis https://doi.org/10.1002/nbm.4504 Editorial
B0 shimming for in vivo magnetic resonance spectroscopy: Experts' consensus recommendations Christoph Juchem, Cristina Cudalbu, Robin A. de Graaf, Rolf Gruetter, Anke Henning, Hoby P. Hetherington, Vincent O. Boer NMR in Biomedicine 2020;e4350 https://doi.org/10.1002/nbm.4350
Advanced magnetic resonance spectroscopic neuroimaging: Experts' consensus recommendations Andrew A. Maudsley, Ovidiu C. Andronesi, Peter B. Barker, Alberto Bizzi, Wolfgang Bogner, Anke Henning, Sarah J. Nelson, Stefan Posse, Dikoma C. Shungu, Brian J. Soher NMR in Biomedicine 2020;e4309 https://doi.org/10.1002/nbm.4309
Motion correction methods for MRS: experts' consensus recommendations Ovidiu C. Andronesi, Pallab K. Bhattacharyya, Wolfgang Bogner, In‐Young Choi, Aaron T. Hess, Phil Lee, Ernesta M. Meintjes, M. Dylan Tisdall, Maxim Zaitzev, André van der Kouwe NMR in Biomedicine 2020;e4364 https://doi.org/10.1002/nbm.4364
Preprocessing, analysis and quantification in single‐voxel magnetic resonance spectroscopy: experts' consensus recommendations Jamie Near Ashley D. Harris Christoph Juchem Roland Kreis Małgorzata Marjańska Gülin Öz Johannes Slotboom Martin Wilson Charles Gasparovic NMR in Biomedicine 2020;e4257 https://doi.org/10.1002/nbm.4257
Proton magnetic resonance spectroscopy in skeletal muscle: Experts' consensus recommendations Martin Krššák, Lucas Lindeboom, Vera Schrauwen‐Hinderling, Lidia S. Szczepaniak, Wim Derave, Jesper Lundbom, Douglas Befroy, Fritz Schick, Jürgen Machann, Roland Kreis, Chris Boesch NMR in Biomedicine 2020;e4266 https://doi.org/10.1002/nbm.4266
31P magnetic resonance spectroscopy in skeletal muscle: Experts' consensus recommendations Martin Meyerspeer, Chris Boesch, Donnie Cameron, Monika Dezortová, Sean C. Forbes, Arend Heerschap, Jeroen A.L. Jeneson, Hermien E. Kan, Jane Kent, Gwenaël Layec, Jeanine J. Prompers, Harmen Reyngoudt, Alison Sleigh, Ladislav Valkovič, Graham J. Kemp, Experts' Working Group on 31P MR Spectroscopy of Skeletal Muscle NMR in Biomedicine 2020;e4246 https://doi.org/10.1002/nbm.4246
Other review papers in the special issue now online
Terminology and concepts for the characterization of in vivo MR spectroscopy methods and MR spectra: Background and experts' consensus recommendations Roland Kreis, Vincent Boer, In‐Young Choi, Cristina Cudalbu, Robin A. de Graaf, Charles Gasparovic,,Arend Heerschap, Martin Krššák, Bernard Lanz, Andrew A. Maudsley, Martin Meyerspeer, Jamie Near, Gülin Öz, Stefan Posse, Johannes Slotboom, Melissa Terpstra, Ivan Tkáč, Martin Wilson, Wolfgang Bogner, Experts' Working Group on Terminology for MR Spectroscopy NMR in Biomedicine 2020;e4347 https://doi.org/10.1002/nbm.4347
Spectral editing in 1H magnetic resonance spectroscopy: Experts' consensus recommendations In‐Young Choi, Ovidiu C. Andronesi, Peter Barker, Wolfgang Bogner, Richard A. E. Edden, Lana G. Kaiser, Phil Lee, Małgorzata Marjańska, Melissa Terpstra, Robin A. de Graaf NMR in Biomedicine 2020;e4411 https://doi.org/10.1002/nbm.4411
Magnetic resonance spectroscopy in the rodent brain: Experts' consensus recommendations Bernard Lanz, Alireza Abaei, Olivier Braissant, In‐Young Choi, Cristina Cudalbu, Pierre‐Gilles Henry, Rolf Gruetter, Firat Kara, Kejal Kantarci, Phil Lee, Norbert W. Lutz, Małgorzata Marjańska, Vladimír Mlynárik, Volker Rasche, Lijing Xin, Julien Valette, the Experts' Working Group on Magnetic resonance spectroscopy in the rodent brain NMR in Biomedicine 2020;e4325 https://doi.org/10.1002/nbm.4325
Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: Experts' consensus recommendations Gülin Öz Dinesh K. Deelchand Jannie P. Wijnen Vladimír Mlynárik Lijing Xin Ralf Mekle Ralph Noeske Tom W.J. Scheenen Ivan Tkáč the Experts' Working Group on Advanced Single Voxel 1H MRS https://doi.org/10.1002/nbm.4236 NMR in Biomedicine. 2020;e4236
Additional review and research papers
Across‐vendor standardization of semi‐LASER for single‐voxel MRS at 3T Dinesh K. Deelchand Adam Berrington Ralph Noeske James M. Joers Arvin Arani Joseph Gillen Michael Schär Jon‐Fredrik Nielsen Scott Peltier Navid Seraji‐Bozorgzad Karl Landheer Christoph Juchem Brian J. Soher Douglas C. Noll Kejal Kantarci Eva M. Ratai Thomas H. Mareci Peter B. Barker Gülin Öz NMR in Biomedicine.2019;e4218 https://doi.org/10.1002/nbm.4218
Multi-vendor standardized sequence for edited magnetic resonance spectroscopy Muhammad G.Saleh Daniel Rimbault Mark Mikkelsen Georg Oeltzschner Anna M.Wang Dengrong Jiang Ali Alhamud Jamie Near Michael Schär Ralph Noeske James B.Murdoch Lars Ersland Alexander R.Craven Gerard Eric Dwyer Eli Renate Grüner LiPann Sinyeob Ahnn Richard A.E.Edden Neuroimage 189 (2019) 425-431 https://doi.org/10.1016/j.neuroimage.2019.01.056
Big GABA II: Water-referenced edited MR spectroscopy at 25 research sites MarkMikkelsen Daniel L.Rimbault Peter B.Barker Pallab K.Bhattacharyya Maiken K.Brix Pieter F.Buur Kim M.Cecil Kimberly L.Chanab David Y.-T.Chen Alexander R.Craven KoenCuypers MichaelDacko Niall W.Duncan UlrikeDydak David A.Edmondson GabrieleEnde LarsErsland Megan A.Forbes FeiGao IanGreenhouse Ashley D.Harris NayingHe StefanieHeba NigelHoggard Tun-WeiHsu Jacobus F.A.Jansen AlayarKangarlu ThomasLang R. MarcLebel YanLi Chien-Yuan E.Lin Jy-KangLiou Jiing-FengLirng FengLiu Joanna R.Long RuoyunMaq CelineMaes MartaMoreno-Ortega Scott O.Murray SeanNoah RalphNoeske Michael D.Noseworthy GeorgOeltzschner Eric C.Porges James J.Prisciandaro Nicolaas A.J.Puts Timothy P.L.Roberts MarkusSack NapaponSailasuta Muhammad G.Saleh Michael-PaulSchallmo NicholasSimard DiederickStoffers Stephan P.Swinnen MartinTegenthoff PeterTruong GuangbinWang Iain D.Wilkinson Hans-JörgWittsack Adam J.Woods HongminXu FuhuaYan ChenchengZhang VadimZipunnikov Helge J.Zöllner Richard A.E.Edden https://doi.org/10.1016/j.neuroimage.2019.02.059
Validation of in vivo MRS measures of metabolite concentrations in the human brain Elvisha Dhamala Ines Abdelkefi Mavesa Nguyen T. Jay Hennessy Hélène Nadeau Jamie Near NMR in Biomedicine. 2019;32:4058 https://doi.org/10.1002/nbm.4058
In vivo diffusion‐weighted MRS using semi‐LASER in the human brain at 3 T: Methodological aspects and clinical feasibility Guglielmo Genovese, Małgorzata Marjańska, Edward J. Auerbach, Lydia Yahia Cherif, Itamar Ronen, Stéphane Lehéricy, Francesca Branzoli NMR in Biomedicine 2020;e4206 https://doi.org/10.1002/nbm.4206
Correcting frequency and phase offsets in MRS data using robust spectral registrationMark Mikkelsen, Sofie Tapper, Jamie Near, Stewart H. Mostofsky, Nicolaas A. J. Puts, Richard A. E. Edden NMR in Biomedicine 2020;e4368 https://doi.org/10.1002/nbm.4368
In vivo Glx and Glu measurements from GABA‐edited MRS at 3 TTiffany Bell, Elodie S. Boudes, Rachelle S. Loo, Gareth J. Barker, David J. Lythgoe, Richard A.E. Edden, R. Marc Lebel, Martin Wilson, Ashley D. Harris NMR in Biomedicine 2020;e4245 https://doi.org/10.1002/nbm.4245
Influence of fitting approaches in LCModel on MRS quantification focusing on age‐specific macromolecules and the spline baseline Małgorzata Marjańska, Melissa Terpstra NMR in Biomedicine 2019;e4197 https://doi.org/10.1002/nbm.4197
Localized MRS reliability of in vivo glutamate at 3 T in shortened scan times: A feasibility study – Efforts to improve rigor and reproducibility Randy P. Auerbach, Diego A. Pizzagalli NMR in Biomedicine 2019;e4093 https://doi.org/10.1002/nbm.4093
Insights into brain microstructure from in vivo DW-MRS Marco Palombo, Noam Shemesh, Itamar Ronen, Julien Valette Neuroimage Volume 182, 15 November 2018, Pages 97-116 https://doi.org/10.1016/j.neuroimage.2017.11.028
Several new publications here on advanced editing techniques - http://www.gabamrs.com/
Voxel placement publication
AutoVOI: real‐time automatic prescription of volume‐of‐interest for single voxel spectroscopy Young Woo Park Dinesh K. Deelchand James M. Joers Brian Hanna Adam Berrington Joseph S. Gillen Kejal Kantarci Brian J.Soher Peter B. Barker HyunWook Park Gülin Öz Christophe Lenglet Magn Reson MEd. 2018;80:1787-1798 https://doi.org/10.1002/mrm.27203
New open-source processing package currently under evaluation -
Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data Georg Oeltzschner Helge J.Zöllner Steve C.N.Hui Mark Mikkelsen Muhammad G.Saleh Sofie Tapper Richard A.E. Edden Journal of Neuroscience Methods Volume 343, 1 September 2020, 108827 https://doi.org/10.1016/j.jneumeth.2020.108827
Presentation at 2020 ENC Meeting - Accelerated MR spectroscopic imaging—a review of current and emerging techniques
Accelerated MR spectroscopic imaging—a review of current and emerging techniques Wolfgang Bogner Ricardo Otazo Anke Henning NMR in Biomedicine.2020;e4314 https://doi.org/10.1002/nbm.4314
WIPs Information:
[1] J. Star-Lack et al., In Vivo Lactate Editing with Simultaneous Detection of Choline, Creatine, NAA, and Lipid Singlets at 1.5 T Using PRESS Excitation with Applications to the Study of Brain andHead and Neck Tumors, J Magn Reson, 133: 243 – 254 (1998) https://doi.org/10.1006/jmre.1998.1458
[2] M. Mikkelsen et al., Big GABA: Edited MR spectroscopy at 24 research sites, NeuroImage, 159, 32 – 45, (2017) https://doi.org/10.1016/j.neuroimage.2017.07.021
[3] M.G. Saleh et al., Multi-vendor standardized sequence for edited magnetic resonance spectroscopy, NeuroImage, (2019) https://doi.org/10.1016/j.neuroimage.2019.01.056
[1] G. Öz et al., Short-Echo, Single-Shot, Full-Intensity Proton Magnetic Resonance Spectroscopy for Neurochemical Profiling at 4 T: Validation in the Cerebellum andBrainstem, Magn Reson Med, 65: 901 – 910 (2011) https://doi.org/10.1002/mrm.22708
[2] VO. Boer et al., 7-T 1H MRS with adiabatic refocusing at short TE using radiofrequency focusing with a dualchannel volume transmit coil, NMR Biomed, 24(9), 1038 – 1046, (2011) https://doi.org/10.1002/nbm.1641
[3] TW. Scheenen et al., Short Echo Time 1H-MRSI of the Human Brain at 3T With Minimal Chemical Shift Displacement Errors Using Adiabatic Refocusing Pulses, Magn Reson Med, 59: 1 – 6 (2008) https://doi.org/10.1002/mrm.21302
Spectroscopy
For users who are totally new to spectroscopy a good basic reference is MRI From Picture to Proton Donald W. McRobbie, Elizabeth A. Moore, Martin J. Graves, and Martin Prince, Cambridge University Press, Second Edition 2007, Chapter 15 (It's not just squiggles: in vivo spectroscopy). This chapter describes spectroscopy from the clinical side, but covers the basics for both data acquisition and processing.
Books:
In Vivo NMR Spectroscopy, 2nd Edition, Author: Robin de Graaf, Wiley Online Library
A recently published book Magnetic Resonance Spectroscopy - Tools for Neuroscience Research and Emerging Clinical Applications
Collecting Data - available sequences and protocols, how-to guides
Current spectroscopy sequences include methods for multi-metabolite data acquisition (Optimized-PRESS [4], [5], [6]) and semi-LASER [9], [10], [11], [12], [13]) and for edited GABA (gamma-Aminobutyric acid) specific data acquisition (MEGA-PRESS [1])
This protocol (spectro-protocol-1 located in the CNI Other tab on the scanner) contains the optimized-PRESS and sLaser sequences for data collection for all metabolites
optimized-PRESS
[4] Webb PG, Sailasuta N, Kohler SJ, Raidy T, Moats RA, Hurd R (1994) Automated single voxel proton MRS: technical development and multisite verification, Magnetic Resonance in Medicine 31(4):365-373 https://doi.org/10.1002/mrm.1910310404
[5] Bodenhausen G, Freeman R, Turner DL (1977) Suppression of artifacts in two dimensional J spectroscopy, Journal of Magnetic Resonance Imaging 27:511-514 https://doi.org/10.1016/0022-2364(77)90016-6
[6] Tran TK, Vigneron DB, Sailasuta N, Tropp J, Le Roux P, Kurhanewicz J, Nelson S, Hurd R (2000) Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer, Magnetic Resonance in Medicine 43(1):23-33. https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291522-2594%28200001%2943%3A1%3C23%3A%3AAID-MRM4%3E3.0.CO%3B2-E
 |
 |
 |
s-Laser
High field (3T) and ultra-high field (7T) are ideal field strength for MR spectroscopy due to the higher spectral resolution and higher signal that can be achieved. But with these advantages comes a higher B1-inhomogeneity and larger Chemical Shift Displacement Error (CSDE). Semi-LASER is a double spin-echo MRS technique like the established PRESS (GE product name Probe-P) technique that uses a slice selective non-adiabatic excitation and two pairs of adiabatic slice selective refocusing pulses for volume selection. The adiabatic behavior of the RF pulses addresses the B1-inhomogeneity problem while the increased bandwidth of these pulses reduces the CSDE.
[9] Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A. (2018) Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses, Magn. Reson. Med. 59(1):1-6 https://doi.org/10.1002/mrm.21302
[10] Oz G, Tkac I. (2011) Short-echo, single-shot, full-intensity proton magnetic resonance spectroscopy for neurochemical profiling at 4 T: validation in the cerebellum and brainstem, Magn. Reson. Med. 65(4):901-910 https://doi.org/10.1002/mrm.22708
[11] Martin Wilson, Ovidiu Andronesi, Peter B. Barker, Robert Bartha, Alberto Bizzi, Patrick J. Bolan, Kevin M. Brindle, In-Young Choi, Cristina Cudalbu, Ulrike Dydak, Uzay E. Emir, Ramon G. Gonzalez, Stephan Gruber, Rolf Gruetter, Rakesh K. Gupta, Arend Heerschap, Anke Henning,Hoby P. Hetherington, Petra S. Huppi, Ralph E. Hurd, Kejal Kantarci, Risto A Kauppinen, Dennis W. J. Klomp, Roland Kreis, Marijn J. Kruiskamp, Martin O. Leach, Alexander P. Lin, Peter R. Luijten, Malgorzata Marjanska, Andrew A. Maudsley, Dieter J. Meyerhoff, Carolyn E. Mountford, Paul G. Mullins, James B. Murdoch, Sarah J. Nelson, Ralph Noeske, Gulin Oz, Julie W. Pan, Andrew C. Peet, Harish Poptani, Stefan Posse, Eva-Maria Ratai, Nouha Salibi, Tom W. J. Scheenen, Ian C. P. Smith, Brian J. Soher, Ivan Tkac, Daniel B. Vigneron, Franklyn A. Howe (2019) Methodological consensus on clinical proton MRS of the brain:Review and recommendations, Magn. Reson. Med. 82:527–550 https://doi.org/10.1002/mrm.27742
[12] Dinesh K. Deelchand, Adam Berrington, Ralph Noeske, James M. Joers, Arvin Arani, Joseph Gillen, Michael Schar, Jon-Fredrik Nielsen, Scott Peltier, Navid Seraji-Bozorgzad, Karl Landheer, Christoph Juchem, Brian Soher, Douglas C. Noll, Kejal Kantarci, Eva M. Ratai, Thomas H. Marecii, Peter B. Barker, Gulin Oz (2019) Across-vendor standardization of semi-LASER for single-voxel MRS at 3T, NMR in Biomedicine. e4218 https://doi.org/10.1002/nbm.4218
[13] Gulin Oz, Dinesh K. Deelchand, Jannie P. Wijnen, Vladimir Mlynarik, Lijing Xin, Ralf Mekle, Ralph Noeske, Tom W.J. Scheenen, Ivan Tkac, the Experts' Working Group on Advanced Single Voxel 1H MRS (2020) Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: Experts' consensus recommendations, NMR in Biomedicine. e4236 https://doi.org/10.1002/nbm.4236
 |
 |
 |
This protocol (spectro-protocol-editing-1 located in the CNI Other tab on the scanner) contains the MEGA-PRESS sequence for data collection for GABA edited data

MEGA-PRESS
[1] Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R (1998) Simultaneous in vivo spectral editing and water suppression, NMR Biomed. 11:266–272 https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291099-1492%28199810%2911%3A6%3C266%3A%3AAID-NBM530%3E3.0.CO%3B2-J
 |
 |
 |
 |
 |
Processing and Analyis of data with Flywheel, how-to guides
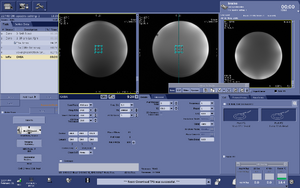
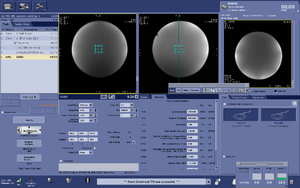
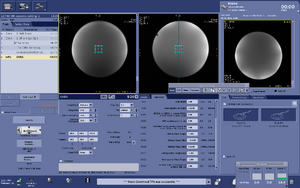
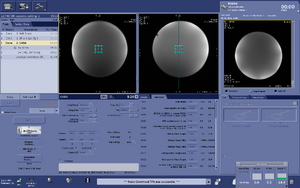
Note: For users using the CNI "nfl" sequence with LCModel data analysis - Users who are in groups that are not themselves Flywheel "Lab" customers will need to download their results from the session tab now (not the analyses tab as was done before and shown in the figure below)
MEGA-PRESS results will continue to be in the analyses tab.

Processing and Analyzing GABA Data - non-Flywheel methods, how-to guides
(1) Analysis of GABA Data collected with the MEGA-PRESS sequence with Gannet
To analyze the data from MEGA-PRESS experiments, we recommend Gannet, a batch-analysis tool for GABA-edited MRS data. The Gannet code and instruction manual can be downloaded from here gabamrs.blogspot.com Additional and newer information can be found here www.gabamrs.com and here www.gabamrs.com/about/ Note that Matlab with Optimization and Statistics Toolboxes must be installed on your computer prior to downloading the Gannet2.0 code.
Note that following change needs to be made to the Gannet code in order to correctly analyze data obtained the CNI MEGA-PRESS sequence gabamrs.blogspot.com (When Gannet goes wrong section)
From gabamrs blogspot -
Solution 2: ON and OFF are incorrectly identified.
If the creatine stripe is correct (red on blue; spectra phased positively) and the difference spectra are still negative, then the issue is the ordering of ON and OFF spectra. The simple solution is to change the MRS_struct.p.onofforder parameter in GannetPreInitialise.m. It is either 'onfirst' or 'offfirst' depending on the acquisition order.
So here's the summary:
"Gannet makes my GABA difference spectra negative".
"Are your creatine signals phased positively?".
If yes, change MRS_struct.p.onofforder; if no, change MRS_struct.p.WaterPositive.
Notes on Spectro Scans on DV26
Prescribing a rotated voxel
At the moment GE’s DV26 software platform release doesn’t support voxel rotation for spectroscopy sequences. A workaround however is available:
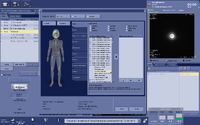
(1) After the 3-plane reconstruction of the T1w images, setup a 3D imaging scan. We saved a template of this 3D scan in the protocol “CNI Example Spectroscopy” and named it as “Voxel prescription”. Add this sequence to your protocol and put it before the MRS scan.
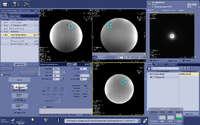
(2) Setup the voxel prescription scan. The Scan Plane is set to “Oblique”, FOV and Locs per Slab are set as close to the voxel size of the spectro scan as possible. Prescribe the 3D box, place it to the desired location and rotate to the right angle. Because the box is usually bigger than the voxel size you want to prescribe, the coverage you see in this step is not accurate. It’s crucial to get the correct orientation of the box. Save Rx.
(3) Setup the MRS scan. Copy Rx from the voxel prescription scan. By default the Mode Filter in the Copy Rx is set to “MRS”, you need to change it to “All” or “3D” in order to see the voxel prescription scan in the Copy Rx list. Select it and accept.
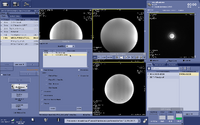
(4) Adjust voxel location and size. You can move the box around and change its size on the graphic interface, or set its coordinates and size by setting the Center and Length in X, Y, Z. If you want to adjust the orientation of the voxel, you need to go back to the 3D voxel prescription scan, and repeat step 2-4 again. After you finish setting up the MRS scan, Save Rx and proceed with scanning.

Extracting voxel prescription information retrospectively
The prescription information of the voxel of an MRS scan is saved in the header of the raw data (p-file). Specifically, the x, y, z dimensions of the voxel are stored in header fields rdb_hdr_image.user8, rdb_hdr_image.user9, rdb_hdr_image.user10; the x, y, z locations of the center of the voxel are stored in header fields rdb_hdr_image.user11, rdb_hdr_image.user12, rdb_hdr_image.user13, in RAS coordinates (e.g. if the voxel is at L26.0 then rdb_hdr_image.user11 will record -26.0). The unit is mm.
To look for this information on Flywheel, open the information window of the p-file, and search for op_user_8, op_user_9, op_user_10, etc.