GE Processing
The GE processing pipeline includes a variety of decisions that advanced users need to know. This page describes some of what we have learned about this pipeline and how to control these decisions. It is our view that each of these decisions should be made very clear to users by GE. Sometimes they are, sometimes not so much.
Return to MR_Protocols
To-do list
Here are various items that are on Kendrick's radar. (Others should feel free to add items.) Higher-priority items are listed towards the top.
- FIELD INHOMOGENEITY CAUSED BY COIL? There is some suspicion that the 32-ch coil causes field inhomogenity near the top on coronal slices. Kendrick will be testing various coils with respect to this issue. The specific reason that the inhomogenity is bad is that if you do a fieldmap correction, the field inhomogeneity can cause bright pixels to be placed in bad locations outside the brain. To avoid this, you could choose your phase encode direction judiciously. For example, with coronal slices, if you choose phase-encode left-right, then the crazy field region is above the head (in the scalp) and won't really have an impact since there are not many bright pixels to the left and right of the top of the scalp.
- FAT/WATER DIFFERENCE. We need to measure the fat/water frequency difference to optimize the TE values used in the spiral fieldmap.
- CAN WE INTRODUCE GRADWARP TO SPIRAL? GE's sequences use gradwarp to fix distortions due to gradient nonlinearities. To ensure consistency, the spiral sequence should also have the gradwarp correction.
- GRADWARP. Can we find out how gradwarp works?
- CAN THE EPI SEQUENCE BE TRIGGERED? I.e., can the stimulus computer send an RF pulse to trigger the EPI sequence? Yes. Use the type-in PSD cni_epi. During prescription, have a look on the Advanced tab and set the triggering option to 1. 0 means "don't wait for a trigger pulse" and 2 means each volume waits for a cardiac trigger pulse.
- SLICE MULTIPLEXING. What's the ETA on this?
Interleave

In the GE world, there are three meanings to the term "interleaved", depending on where you interact with the protocol:
- The interleaved (vs. sequential) radio button in the EPI "Multi-phase" tab means acquire all the slices then repeat. In this context, "sequential" means acquire all time frames for a slice and then repeat for the next slice. For fMRI, you always want this to be set to interleaved.
- interleaved can also mean lines of k-space acquisition are interleaved for multishot EPI.
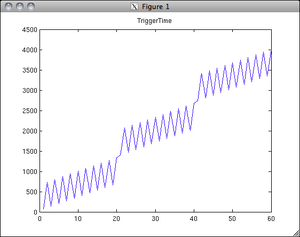
- interleaved can also mean slices are acquired odd first, then even. Kendrick has confirmed that the GE EPI sequence uses interleaved slice ordering by default. It isn't obvious how to change this using the GE interface, but interleaved is generally preferred anyway. Just be sure to take this into account when doing slice timing correction. You can use the "TriggerTime" field in the DICOM head to confirm the slice acquisition timing. (Kendrick also verified that the time points at which slices are acquired are equally spaced within the TR (e.g. see the figure).)
So, for our BOLD EPI sequence, the slice order is odd, even. E.g., for a 20-slice prescription, the slice order would be [1 3 5 7 9 2 4 6 8 10]. Slice 1 is the first slice in your NIFTI file. For an axial prescription, this slice might be either the most inferior or the most superior slice, depending on the slice order defined by the prescription. (The slice numbers appear in the graphic Rx dialog on the scanner console.) We are confident that the NIFTI files generated by the CNI NIMS system contain the correct slice order and necessary NIFTI header information to accurately indicate the slice order. However, if you have questions about this, contact the CNI Research Director.
Gradwarp
- By default, the scanner applies a correction for nonlinearities in the gradients, called gradwarp. Presumably, the correction will make your images more spatially accurate. It is not clear exactly what is the nature of the correction.
- Currently, Atsushi's spiral sequence does not take the gradwarp into account; thus, spiral images may be (slightly) misregistered with respect to images from stock GE sequences.
- Based on inspection of data, it appears that gradwarp corrects in 3D for the 3D T1 Bravo sequence. But perhaps the correction is only 2D for 2D sequences?
- It appears that the gradient warping step is the very final step applied to the data (i.e., after calculating the magnitude, phase, real, and imaginary images, the gradient warping induces one final interpolation of the images). If you are interested in the phase data, you should compute the phase yourself from the gradwarped real and imaginary images instead of using the gradwarped phase image (which has interpolation problems).
- Perhaps we could make use of the Gradient Non-linearity Unwarping Tool; also see the Neuroimage article by Jovicich, "Reliability in multi-site structural MRI studies: Effects of gradient non-linearity correction on phantom and human data".
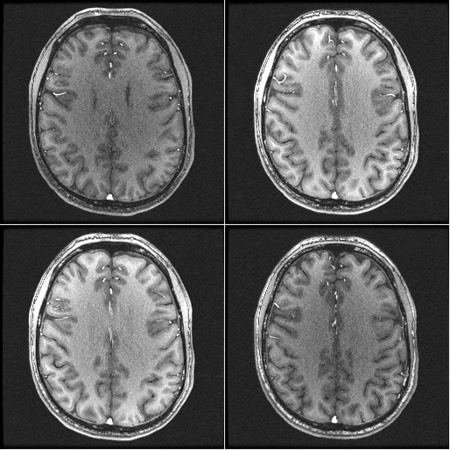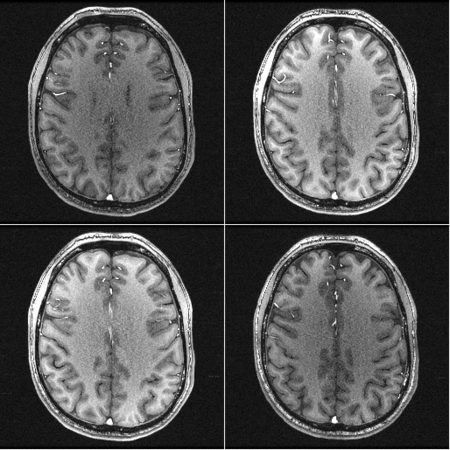
This is an example of the effect of gradwarp:
There are two frames. The frame with a darker background and bigger brain is a version with the gradwarp correction turned OFF. The other frame has the gradwarp correction turned ON (which is the default);
"Silent" Partial Fourier
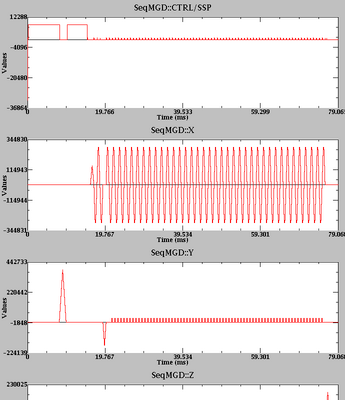
- In EPI, if you increase the acquisition time too much (e.g. by having a large matrix size or a short TE), the pulse sequence will be unable to acquire the requisite half of k-space before the TE value. When this happens, the scanner silently decides to simply drop that half of k-space and acquire a default of 8 lines of k-space for that half. This is potentially quite bad, as you lose a lot of SNR, since there will be wasted dead time after the RF pulse up until the beginning of the 8 lines. Moreover, partial acquisitions may result in difficulties in the reconstruction process and thus image artifacts (I have noticed these when scanning phantoms). The safest way to see whether the silent dropping of k-space happens is to check the plotter (to do that, start scanning, pause the scan, and then run plotter from a command window). An easier (but unverified) way is to monitor the Relative SNR number in the console. It appears that once you go over the limit, the relative SNR jumps way down.
- There is weird buggy behavior in the console concerning the relative SNR number. If you violate the limit and the number of slices that can be acquired is less than the current number of slices (resulting in # acqs being set to 2), you need to reset the number of slices to a lower number within the limit. Then, the # acqs will go back to 1, as desired.
- Regarding using the relative SNR as a index for whether the silent partial fourier occurs, for some reason, you need at least 20 volumes prescribed. I have found that if you prescribe less volumes, then the big jump in relative SNR does not occur.
- To deal with the "silent" partial Fourier issue, one strategy is to ensure that you always get full coverage of k-space.
- A different strategy is to ensure that you get the maximal number of lines of k-space (not the default 8 lines). It is not clear how to do this (perhaps CV variable num_overscan?)
Here's an example of what happens. This is 70 phase-encode lines:
Everything is OK; a full k-space acquisition is obtained.
This is 72 phase-encode lines:
Still OK.
This is 74 phase-encode lines:
OOPS. Now there is substantial dead time before the TE.
Here's some more illustration of what the problem really amounts to. This is a phantom, using 72 phase-encode lines:
And this is the temporal SNR (red and dark red is good):
This is the same phantom, if you increase the phase-encode lines to 74 (which triggers the silent partial Fourier):
Notice the reconstruction artifacts. And this is the temporal SNR:
Notice it's substantially worse than the temporal SNR of the 72 version.
Fermi filtering
- The reconstruction code applies a Fermi low-pass filter by default(!). To turn this off, set CV var rhfermr to the matrix size and set CV var rhfermw to 1. Unfortunately, it appears that these CV vars are not saved when you save protocols, so you'll have to do this at least once per scan session.
- Apparently, the purpose of Fermi filtering is to reduce Gibbs artifacts (which are caused by truncation of k-space).
Reconstructed image size
- By default, images are reconstructed at matrix sizes that are powers of two (presumably for speed). However, this wastes disk and memory space. To change the reconstructed image size, set CV variables rhrcxres,rhrcyres,rhimsize. For example, if I acquire a 70 x 70 matrix size, I would set rhrcxres=70, rhrcyres=70, rhimsize=70, and then I would get 70x70 images instead of 128x128. Unfortunately, it appears that these CV vars are not saved when you save protocols, so you'll have to do this at least once per scan session.
- But be careful --- it appears that one you set these CV vars, they will no longer default to the next highest power of two if you change parameters of the sequence. For example, in one instance, I was increasing the matrix size but forgot to change the CV vars, and the resulting reconstructed images were totally corrupted. It is important to visually inspect the data in the console as they are acquired in order to verify sanity!
Precise values
- The GUI tends to round values that you enter. For example, if you enter 1605.242 for the TR, the GUI will round the value displayed to 1605.2, but the value that actually will get used is the precise one. You can check this via the CV vars (e.g. optr), which you can take to be the actual value used.
- If you enter in a precise value, and copy and paste the entire sequence, usually I find that the precise value is also copied.
- If you enter in a precise value, and then save the protocol, I have found that (at least in some cases) the wrong, rounded value is saved. You should re-enter the precise value every time you load in the saved protocol.
- If you have a precise FOV size (e.g. 14.24 cm FOV) entered, and you copy that slice prescription to another sequence, I have found that the rounded FOV is copied (e.g. 14.2 cm FOV). You should re-enter the value to get it right.
Phase data
- By default, only magnitude images are reconstructed and saved by the scanner. You can set the CV variable rhrcctrl in order to save phase-related images in addition to the magnitude images. It appears that rhrcctrl should be an integer between 0 and 15, and reflects a 4-bit mask where the ordering of bits (from high to low) are imaginary, real, phase, magnitude. For example, rhrcctrl=13 means to save imaginary, real, and magnitude images.
- In the DICOM files outputted by the scanner, the scanner cycles through the various versions of each slice before proceeding to the next slice. The order appears to be magnitude, phase, real, and then imaginary.
- I have found that if you try to write out all four versions of the images, the reconstruction time becomes a real issue to worry about --- if you attempt to start the next EPI run before the scanner completes the reconstruction of the data from the previous EPI run, it will pause the new EPI run somewhere in the middle (presumably when its cache fills up or something). This is really bad, so you should wait until the previous run completely finishes its reconstruction. To speed things up, consider writing only the versions of the images that you need.
CV variables
Many variables that control scanner behavior are not accessible via the usual GUI interface. To access these variables, press Save Rx and then Research->Download. Then, select Display CVs.
Here is a list of CV vars that Kendrick has run across. Atsushi might know about more of them.
YOU MAY VERY WELL WANT TO USE THESE: map_deltaf [frequency for echo time difference (for Atsushi's spiral fieldmap)] rhfermr [Fermi radius (in matrix units I think)] rhfermw [Fermi width (in matrix units I think)] rhrcxres [transform size (x)] rhrcyres [transform size (y)] rhimsize [image size] pepolar [phase encoding polarity (direction)]
YOU PROBABLY DON'T WANT TO USE THESE: opslquant [number of slices] optr [TR] opfphases [number of TRs] opte [TE] opflip [flip angle] opfov [field-of-view] opxres [frequency-encode resolution] opyres [phase-encode resolution] opslthick [slice thickness] opphasefov [fraction of the FOV in the phase-encode direction] avminte [minimum TE, same as in GUI] opnex [number of excitations] nograd [no gradwarp, default is 0] rhferme [Fermi eccentricity??] rhrcctrl [controls what kinds of images to save (see "Phase data" section above)]
UNKNOWN/UNTESTED: nframes [number of frames?] opti [?] num_overscan [default is 0?] rhmethod [default is 1?] esp [echo spacing?] autolock [autolock raw files, default is 0] rhexecctrl [?]
Fieldmaps
Kendrick's Notes on Fieldmaps
One of the major problems that affects EPI data is spatial distortion. It is possible to correct for distortions in EPI data posthoc using fieldmap measurements. Kendrick has developed some MATLAB code that handles spatial distortion and other pre-processing procedures relevant to EPI fMRI data.
The process can be divided into two basic parts. The first part is getting the fieldmap acquisition right. The second part is using the code to process the EPI and fieldmap data.
Fieldmap strategy
The higher-order shim (HOShim) is quite important and massages the field to be as homogenous as possible. (It's best to get the field right up front instead of relying on postprocessing!). In the HOShim, you should probably prescribe an ellipse that is liberal, i.e. covers all parts of the brain that are within your slice prescription. Covering some air outside the brain is fine. After the HOShim, make sure the Shim setting is OFF on all subsequent scans.
Normally, a pre-scan is run before each GE sequence. One of the things that the pre-scan does is to re-measure the peak frequency corresponding to water. This in effect compensates for drift in the overall magnetic field strength (which may be due to heating which in turn is dependent on what pulse sequences you run). But change in the magnetic field is exactly what we are trying to track using fieldmaps. So, some fancy footwork is necessary to avoid re-measuring the peak frequency.
The strategy that Kendrick likes is this:
- After completing the localizer, ASSET calibration, higher-order shim, and in-planes, we are ready to proceed to the actual functional data.
- Set up the EPI sequence and prescribe only one volume. Then hit SCAN as usual. This will trigger the pre-scan and acquire one volume. We want to trigger the pre-scan because the pre-scan includes other calibration routines that reduce ghosting, etc.
- Then, proceed to collect your actual data by sandwiching EPI runs within fieldmap acquisitions, like F1 / E1 / F2 / E2 / F3 / E3 / F4. This ensures that you have a good estimate of the field throughout all of the EPI data.
- Importantly, when starting each EPI and each fieldmap, click Save Rx, then click Research->Download, then Manual Prep Scan. This will open up a window. Don't do anything and just click DONE. Then click Prep Scan. Then press the green button on the console keyboard to start the scan. By using this procedure to start the scan, we avoid the pre-scan and therefore avoid the re-calibration of the peak frequency.
If for some reason, later in the session, you decide to acquire sequences other than fieldmaps and EPIs, you should allow the pre-scan to run as usual (by just hitting the SCAN button to scan). Of course, this will change the global constant describing the field strength.
Also, if you for some reason change parameters of the EPI sequence (such as TR, TE, phase direction), you need to allow the pre-scan to run so that proper calibration can be done. However, certain EPI parameters do not necessitate a pre-scan, like number of volumes to acquire and the phase-encode polarity (which is controlled through the CV variable pepolar).
Admittedly, sandwiching every EPI run between fieldmaps is an aggressive (and paranoid) strategy --- each fieldmap takes about a minute to acquire. If you like, it may be okay to space the fieldmaps more sparsely, e.g. F1 / E1 / E2 / F2 / E3 / E4 / F3, depending on how long your EPI runs are. Of course, this comes at the expense of having sub-optimal tracking of the field.
As an even less aggressive strategy, it is also conceivable to acquire only a single fieldmap (indeed, it appears that that's what most laboratories do, if anything at all) and also allow each EPI run to do the pre-scan (which avoids the cumbersome Manual Prep Scan procedure). The hope here is that the bulk of the drift in the magnetic field can be captured by just a change in a global constant. To a first approximation, it does appear that the overall DC in the field is the biggest effect, but in data that I have looked at, there are additional components that can't be described by just a constant. Moreover, using a single fieldmap cannot compensate for changes in the field within individual EPI runs.
Spiral fieldmap acquisition
Atsushi has developed a spiral-based sequence for measuring fieldmaps, and the quality is quite good. Here are the parameters that Kendrick recommends for optimal quality:
- First, set slice thickness.
- Then copy slices from the EPI (or in-plane) sequence.
- 16-shot (16 spiral interleaves). The purpose is to minimize the readout time so as to minimize distortion and dropout (in combination with an appropriately short TE).
- 700 ms TR. You could try to reduce this value to speed up acquisition. If the other parameters necessitate a longer TR, the sequence will silently use a longer TR and the scanner will scan past the on-screen time countdown. This is okay, just wait a few seconds.
- 54° flip. This is the Ernst angle for the 700 ms TR (assuming a T1 constant of 1333.33 ms).
- no SAT. In regions of bad field inhomogeneity, water will be off-resonance. So if you were to use a fat saturation pulse, you could destroy the water signal in these regions. That is bad. So, leave SAT off.
- 256 x 256. This is a fairly high matrix size which will ensure high-resolution, high-quality field maps.
- 2 NEX. Get two repetitions to ensure that the fieldmaps will have high SNR. If time is a consideration, you might try going down to 1 NEX.
- 24.0 FOV. This is a large FOV to ensure that the entire head is completely covered in the in-plane dimensions. You could reduce, but be careful.
- 6 dummies to ensure we reach a steady-state.
- 4.545 ms TE. This is 1/440Hz * 2. It is unclear whether saved protocols will save the precise TE value. So it is recommended that you manually type in 4.545 in each scan session. (After typing it in, copying and pasting that sequence does preserve the precise value.)
- map_deltaf 440. You must edit this CV variable after saving and downloading the spiral sequence. (The purpose of the TE and map_deltaf values is to choose echo times that are matched to the difference between the precession frequencies of water and fat, so that the resulting fieldmaps will have minimal artifacts.) (UPDATE: This is now the default setting for the map_deltaf, so actually, you don't have to go in and edit the CV variable.)
- Note: it is very important that you get the TE and map_deltaf values right! Otherwise, there will be fat-related artifacts in your fieldmaps.
- You will have to transfer the raw fieldmap data yourself (they do not get converted to DICOM format like data from the stock GE sequences). The files are named like P39936.7.
Pre-processing code
See preprocessfmri for details on how to process EPI data using fieldmaps. Note that you need to know the number of phase encode lines (typically your image size along the phase encode dimension, divided by the ASSET acceleration factor) and the phase encode dwell time (which should be stored in the DICOM field (0043,102c): the time per phase-encode line in microseconds).
Using FSL to process CNI fieldmaps
The ...fieldmap.nii.gz file that we save in NIMS is what the FSL folks call "A single, real fieldmap image (showing the field inhomogeneity in each voxel)". (NOTE: for older datasets in NIMS, this file uses the 'B0' suffix rather than 'fieldmap'.) The other nifti associated with a fieldmap scan is a magnitude image that can be used for masking, etc.
The fieldmap image is in units of Hz. FSL likes radians/s, so you can convert it with fslmaths:
fslmaths 8860_6_1fieldmap.nii.gz -mul 6.28 fieldmap_rads
Then jump straight to step 5 on the FSL FUGUE guide. Although you may want to mask the fieldmap first. E.g.:
bet 8860_6_1.nii.gz fieldmap -m -n fslmaths fieldmap_rads.nii.gz -mul fieldmap_mask.nii.gz fieldmap_rads fugue --loadfmap=fieldmap_rads --despike --savefmap=fieldmap_rads fugue --loadfmap=fieldmap_rads -m --savefmap=fieldmap_rads fugue --loadfmap=fieldmap_rads -s 1 --savefmap=fieldmap_rads
Then you can use FUGUE command-line tool to unwarp the EPI image. E.g.:
fugue -i epi --dwell=dwelltime --loadfmap=fieldmap -u result
where dwelltime is the same as the effective echo spacing. You can find the effective echo spacing from the header of your epi file. E.g., using fslhd, look at 'descrip':
fslhd 8860_5_1.nii.gz | grep 'descrip'
which produces:
descrip te=30.00;ti=0;fa=77;ec=0.4800;acq=[80,80];mt=0;rp=2.0;
showing that the echo spacing ('ec') is 0.48 milliseconds. The phase encoding dimension is usually y for EPI acquisitions. Note that, as the FSL docs say, "your operator can tell you whether the unwarp direction, or phase encode direction, is x, y or z, but whether it is + or - needs to be determined, currently, by trial and error..." However, we think it's usually + for data acquired here.